Abstract
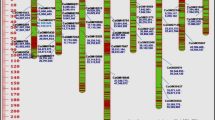
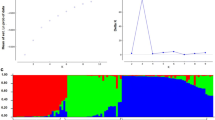
Biodiversity information in available germplasm is very useful for the success of any breeding program. To establish genetic diversity among 44 genotypes of chickpea comprising cultigen, landraces, internationally developed improved lines and wild relatives, genetic distances were evaluated using 19 simple sequence repeat markers with 100 marker loci. Estimation of the number of alleles per locus (n a), the effective allele number (n e), and Wright fixation index F were 6.25, 3.67, and 0.44, respectively. Polymorphism information content values ranged from 0.84 (locus NCPGR6 and TA135) to 0.44 (locus NCPGR7) with an average of 0.68. Dice’s coefficient similarity matrix for studied chickpea genotypes varied from 0.07 to 1.0 indicating a broader genetic base among genotypes studied. The highest similarity, 1.0, was observed between genotypes Sel 96TH11484 and Sel 96TH11485; while, the lowest, 0.07, was observed between genotypes Sel 95TH1716 and Azad. Based on the UPGMA clustering method, all genotypes were clustered in eight groups, which indicated the probable origin and region similarity of landraces and local Iran landraces over the other cultivars and wild species. It also represents a wide diversity among available germplasm. Analysis of molecular variance revealed that 41% of the total variance was due to differences among groups while 59% was due to differences within groups. The results of principal coordinate analysis approximately corresponded to those obtained through cluster analysis. Genetic variation detected in this study can be useful for selective breeding for specific traits and in enhancing the genetic base of breeding programs.



Similar content being viewed by others
References
Agharkar SP (1991) Medicinal plants of Bombay presidency. Scientific, Jodhpur, pp 62–63
Ahmad F (1999) Random amplified polymorphic DNA (RAPD) analysis reveals genetic relationships among the annual Cicer species. Theor Appl Genet 98:657–663. doi:10.1007/s001220051117
Ahmad F, Slinkard AE (1992) Genetic relationships in the genus Cicer L. as revealed by polyacrylamide gel electrophoresis of seed storage proteins. Theor Appl Genet 84:688–692. doi:10.1007/BF00224169
Anderson JA, Churchill GA, Autrique JE, Tanksley SD, Sorrells ME (1992) Optimizing parental selection for genetic linkage maps. Genome 36:181–186. doi:10.1139/g93-024
Arumuganathan K, Earle ED (1991) Nuclear DNA content of some important plant species. Plant Mol Biol Rep 9:208–218
Berger J, Abbo S, Turner N (2003) Ecogeography of annual wild Cicer species: the poor state of the world collection. Crop Sci 43:1076–1090
Bourguiba H, Krichen L, Audergon J-M, Khadari B, Trifi-Farah N (2010) Impact of mapped SSR markers on the genetic diversity of apricot (Prunus armeniaca L.) in Tunisia. Plant Mol Biol Rep 28:578–587
Charles MT, Dominique R, Kumar J, Dangi OP (2002) A preliminary study of the functional properties of chickpea leaves. In: Annual Meeting of the Canadian Society of Food and Nutrition, May 2002, Edmonton, Alberta, Canada pp. 89–96
Choudhary S, Sethy NK, Shokeen B, Bhatia S (2006) Development of sequence-tagged microsatellite site markers for chickpea (Cicer arietinum L.). Mol Ecol Notes 6:93–95. doi:10.1111/j.1471-8286.2005.01150.x
Choumane W, Winter P, Weigand F, Kahl G (2000) Conservation and variability of sequence-tagged microsatellite sites (STMSs) from chickpea (Cicer aerietinum L.) within the genus Cicer. Theor Appl Genet 101:269–278. doi:10.1007/s001220051479
Chowdhury MA, Vandenberg V, Warkentin T (2002) Cultivar identification and genetic relationship among selected breeding lines and cultivars in chickpea (Cicer arietinum L.). Euphytica 127:317–325. doi:10.1023/A:1020366819075
Cingilli H, Altinkut A, Akcin A (2005) The use of microsatellite markers in the annual and perennial Cicer species growing in Turkey. Biologia, Sect Bot 60(1):93–98, RN: SK2005000138
Coram TE, Mantri NL, Ford R, Pang ECK (2007) Functional genomics in chickpea: an emerging frontier for molecular- assisted breeding. Funct Plant Biol 34:861–873. doi:10.1071/FP07169
Dellaporta SL, Wood J, Hicks JB (1983) A plant DNA minipreparation. Plant Mol Biol Rep 1:19–21
Dice LR (1945) Measures of the amount of ecologic association between species. Ecology 26:297–302
Dunna V, Dadlani M, Kumar PA, Panguluri SK (2009) Molecular marker analysis of differentially aged seeds of soybean and safflower. Plant Mol Biol Rep 27(3):282–291
Flandez-Galvez H, Ford R, Pang EC, Taylor PW (2003) An intraspecific linkage map of the chickpea (Cicer arietinum L.) genome based on sequence tagged microsatellite site and resistance gene analog markers. Theor Appl Genet 106:1447–1456. doi:10.1007/s00122-003-1199-y
Gomes S, Martins-Lopes P, Lopes J, Guedes-Pinto H (2009) Assessing genetic diversity in Olea europaea L. using ISSR and SSR markers. Plant Mol Biol Rep 27:365–373
Hallauer AR (1999) Temperate maize and heterosis. In: Coors J and Pandey S (Eds.) Genetics and exploitation of heterosis in crops, CIMMYT, Mexico City 17–22 Aug. 1997 ASA, Madison, WI. pp. 353–361.
Hartl DL, Clark AG (1989) Principles of population genetics. Sinauer, Sunderland
He Q, Li XW, Liang GL, Ji K, Guo QG, Yuan WM, Zhou GZ, Chen KS, Weg EW, Gao ZS (2011) Genetic diversity and identity of Chinese loquat cultivars/accessions (Eriobotrya japonica) using apple SSR markers. Plant Mol Biol Rep 29:197–208
Hernández P, de la Rosa R, Rallo L, Dorado G (2001) Development of SCAR markers in olive (Olea europaea) by direct sequencing of RAPD products: applications in olive germplasm evaluation and mapping. Theor Appl Genet 103:788–791. doi:10.1007/s001220100603
Hutokshi KB, Jayashree B, Eshwar K, Crouch JH (2005) Development of ESTs from chickpea roots and their use in diversity analysis of the Cicer genus. BMC Plant Biol 5:16. doi:10.1186/1471-2229-5-16
Huttel B, Winter P, Weising K, Choumane W, Weigand F, Kahl G (1999) Sequence-tagged microsatellite site markers for chickpea (Cicer arietinum L.). Genome 42:210–217. doi:10.1007/s00122-003-1260-x
Imtiaz M, Materne M, Hobson K, van Ginkel MB, Malhotra RS (2008) Molecular genetic diversity and linked resistance to ascochyta blight in Australian chickpea breeding materials and their wild relatives. Aust J Agric Res 59:554–560. doi:10.1071/AR07386
Iruela M, Rubio J, Cubero JI, Gil J, Millan T (2002) Phylogenetic analysis in the genus Cicer and cultivated chickpea using RAPD and ISSR markers. Theor Appl Genet 104:643–651. doi:10.1007/s001220100751
Kazan K, Muehlbauer FJ (1991) Allozyme variation and phylogeny in annual species of Cicer (Leguminosae). Plant Syst Evol 175:11–21. doi:10.1007/BF00942142
Khan R, Khan H, Farhatullah, Harada K (2010) Evaluation of microsatellite markers to discriminate induced mutation lines, hybrid lines and cultigens in chickpea (Cicer arietinum L). AJCS 4(5):301–308
Labdi M, Robertson LD, Singh KB, Charrier A (1996) Genetic diversity and phylogenetic relationships among the annual Cicer species as revealed by isozyme polymorphism. Euphytica 88:181–188. doi:10.1007/BF00023889
Ladizinsky G, Adler A (1976) Genetic relationships among the annual species of Cicer L. Theor Appl Genet 48:197–203. doi:10.1007/BF00527371
Lichtenzveig J, Scheuring C, Dodge J, Abbo S, Zhang HB (2004) Construction of BAC and BIBAC libraries and their applications for generation of SSR markers for genome analysis of chickpea, Cicer arietinum L. Theor Appl Genet 110(3):492–510. doi:10.1007/s00122-004-1857-8
McIntosh GH, Topping DL (2000) Food legumes in human nutrition. In: Knight R (ed) Linking research and marketing opportunities for pulses in the 21st century. Kluwer, Dordrecht, pp 655–666
Millan T, Siddique CHJ, KHM BHK, Gaur PM, Kumar J, Gil J, Kahl G, Winter P (2006) Chickpea molecular breeding: new tools and concepts. Uphitica 147(1–2):81–103. doi:10.1007/s10681-006-4261-4
Nei NM, Li W (1979) Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci 76:5269–5273, PMCID: PMC413122
Nguyen TT, Taylor PWJ, Redden RJ, Ford R (2004) Genetic diversity estimates in Cicer using AFLP analysis. Plant Breed 123:173–179. doi:10.1046/j.1439-0523.2003.00942.x
Pandian A, Ford R, Taylor PWJ (2000) Transferability of sequence tagged microsatellite site (STMS) primers across four major pulses. Plant Mol Biol Report 18:395a–395h. doi:10.1007/BF02825069
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295. doi:10.1111/j.1471-8286.2005.01155.x
Popelka JC, Higgins TJV (2007) Chickpea. In: Pua EC, Davey MR (eds) Biotechnology in agriculture and forestry, Vol. 59 transgenic crops IV. Springer, Berlin, pp 251–262
Powell W, Morgante M, Andre C, Hanafey M, Vogel J, And TS, Rafalski A (1996) The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Mol Breed 2:225–238. doi:10.1007/BF00564200
Rao LS, Usha Rani P, Deshmukh PS, Kumar PA, Panguluri SK (2007) RAPD and ISSR fingerprinting in cultivated chickpea (Cicer arietinum L.) and its wild progenitor Cicer reticulatum Ladizinsky. Genet Resour Crop Evol 54:1235–1244
Ratnaparkhe MB, Gupta VS, Ven Murthy MR , Ranjekar PK (1995) Genetic fingerprinting of pigeon Cajanus cajan (L.) Millsp and its wild relatives using RAPD markers. Theor. Appl. Genet. 91: 893–898. doi:10.1007/BF00223897
Rohlf FJ (1998) NTSYS-pc: Numerical taxonomy and multivariate analysis system, version 2.02. Exter Software, Setauket, New York. http://www.exetersoftware.com/cat/ntsyspc/ntsyspc.html. Accessed 12 Agust 2010
Saeed A, Darvishzadeh R, Hovsepyan H, Asatryan A (2010) Tolerance to freezing stress in Cicer accessions under controlled and field conditions. Afr J Biotechnol 9(18):2618–2626
Sant VJ, Patankar AG, Sarode ND, Mhase LB, Sainani MN, Deshmukh RB, Ranjekar PK, Gupta VS (1999) Potential of DNA markers in detecting divergence and analysis in heterosis in Indian elite chickpea cultivars. Theor Appl Genet 98:1217–1225. doi:10.1007/s001220051187
Senior ML, Murphy JP, Goodman MM, Stuber CW (1998) Utility of SSRs for determining genetic similarities and relationships in maize using an agarose gel system. Crop Sci 38:1088–1098
Serret MD, Udupa SM, Weigand F (1997) Assessment of genetic diversity of cultivated chickpea using microsatellite-derived RFLP markers: implications for origin. Plant Breed 116:573–578. doi:10.1111/j.1439-0523.1997.tb02192.x
Sethy NK, Shokeen B, Bhatia S (2003) Isolation and characterization of sequence-tagged microsatellite sites markers in chickpea (Cicer arietinum L.). Mol. Ecol. Notes 3:428–430. doi:10.1046/j.1471-8286.2003.00472.x
Sethy NK, Choudhary S, Shokeen B, Bhatia S (2006a) Identification of microsatellite markers from Cicer reticulatum: molecular variation and phylogenetic analysis. Theor Appl Genet 112:347–357. doi:10.1007/s00122-005-0135-8
Sethy NK, Shokeen B, Edwards KJ, Bhatia S (2006b) Development of microsatellite markers and analysis of intraspecific genetic variability in chickpea (Cicer arietinum L.). Theor Appl Genet 112:1416–1428. doi:10.1007/s00122-006-0243-0
Sharma PC, Huttel B, Winter P, Kahl G, Gardner RC, Weising K (1995) The potential of microsatellites for hybridization- and polymerase chain reaction-based DNA fingerprinting of chickpea (Cicer arietinum L.) and related species. Electrophoresis 16:1755–1761
Singh KB (1987) Chickpea breeding. In: Saxena MC, Singh KB (eds) The chickpea. CAB, Wallingford, pp 127–162
Stuber CW (1994) Heterosis in plant breeding. Plant Breed Rev 12:227–251
Sudupak M (2004) Inter and intra-species inter simple sequence repeat (ISSR) variation in the genus Cicer. Euphytica 135:229–238. doi:10.1023/B:EUPH.0000014938.02019.f3
Sudupak A, Akkaya S, Kence A (2002) Analysis of genetic relationships among perennial and annual Cicer species growing in Turkey using RAPD markers. Theor Appl Genet 105:1220–1228. doi:10.1007/s00122-002-1060-8
Sun GL, William M, Liu J, Kasha KJ, Pauls KP (2001) Microsatellite and RAPD polymorphisms in Ontario corn hybrids are related to the commercial sources and maturity ratings. Mol Breed 7:13–24. doi:10.1023/A:1009680506508
Taylor PWJ, Ford R (2003) Chickpea. In: Kole C (Ed.) Genome mapping and molecular breeding in plants, Volume 3 Pulses, Sugar and Tuber Crops © Springer-Verlag Berlin Heidelberg
Udupa SM, Baum M (2001) High mutation rate and mutational bias at (TAA)n microsatellite loci in chickpea (Cicer arietinum L.). Mol Gen Genom 265:1097–1103. doi:10.1007/s004380100568
Udupa SM, Robertson LD, Weigand F, Baum M, Kahl G (1999) Allelic variation at (TAA)n microsatellite loci in a world collection of chickpea (Cicer arietinum L.) germplasm. Mol Gen Genet 261:354–363. doi:10.1007/s004380050976
Upadhaya HD, Dwivedi SL, Gowda CLL, Singh S (2007) Identification of diverse germplasm lines for agronomic traits in chickpea (Cicer arietinum L.) core collection for use in crop improvement. Field Crops Res 100:320–326
Upadhyaya HD, Dwivedi SL, Baum M, Varshney RK, Udupa SM, Gowda CLL (2008) Genetic structure, diversity, and allelic richness in composite collection and reference set in chickpea (Cicer arietinum L.). BMC Plant Biol 8:106. doi:10.1186/1471-2229-8-106
Varshney RK, Hoisington DA, Upadhyaya HD, Gaur PM, Nigam SN, Saxena KB, Vadez V, Sethy NK, Bhatia S, Aruna R, Gowda MVC, Singh NK (2007a) Molecular genetics and breeding of grain legume crops for the semi-arid tropics. In Genomics Assisted Crop Improvement, Genomics Applications. In: Varshney RK, Tuberosa R, Springer D (Eds.) Crops.Volume II. The Netherlands. pp. 207–242
Varshney RK, Coyne CJ, Swamy P, Hoisington D (2007b) Molecular identification of genetically distinct accessions in the USDA chickpea core collection. Pisum Genet 39:32–33
Winter P, Pfaff T, Udupa SM, Huttel B, Sharma PC, Sahi S, Arreguin-Espinoza R, Weigand F, Muehlbauer FJ, Kahl G (1999) Characterization and mapping of sequence-tagged microsatellite sites in the chickpea (Cicer arietinum L.) genome. Mol Gen Genet 262:90–101. doi:10.1007/s004380051063
Winter P, Benko-Iseppon AM, Huttel B, Ratnaparkhe M, Tullu A, Sonnante G, Pfaff T, Tekeoglu M, Santara D, Sant VJ, Rajesh PN, Kahl G, Muehlbauer FJ (2000) A linkage map of chickpea (Cicer arietinum L.) genome based on recombinant inbred lines from a C. arietinum and C. reticulatum cross: localization of resistance genes for fusarium wilt races 4 and 5. Theor Appl Genet 101:1155–1163. doi:10.1007/s001220051592
Ye C, Yu Z, Kong F, Wu S, Wang B (2005) R-ISSR as a new tool for genomic fingerprinting, mapping, and gene tagging. Plant Mol Biol Rep 23:167–177
Yeh FC, Yang RC, Boyle T (1999) “POPGENE version 1.31: Microsoft Window- based Freeware for Population Genetic Analysis” available at: http://www.ualberta. ca/∼fyeh. Accessed 21 September 2010
Zhao F, Cai Z, Hu T, Yao H, Wang L, Dong N, Wang B, Ru Z, Zhai W (2010) Genetic analysis and molecular mapping of a novel gene conferring resistance to rice stripe virus. Plant Mol Biol Rep 28:512–518
Acknowledgment
The authors are grateful to the Institute of Biotechnology, Urmia University, Iran for the scientific support of this work.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Saeed, A., Hovsepyan, H., Darvishzadeh, R. et al. Genetic Diversity of Iranian Accessions, Improved Lines of Chickpea (Cicer arietinum L.) and Their Wild Relatives by Using Simple Sequence Repeats. Plant Mol Biol Rep 29, 848–858 (2011). https://doi.org/10.1007/s11105-011-0294-5
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-011-0294-5