Abstract
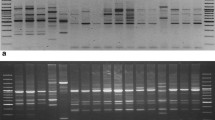
Shisham (Dalbergia sissoo) is one of the most preferred timber tree species of South Asia. Two DNA-based molecular marker techniques, intersimple sequence repeat (ISSR) and random amplified polymorphism DNA (RAPD), were compared to study the genetic diversity in this species. A total of 30 polymorphic primers (15 ISSR and 15 random) were used. Amplification of genomic DNA of 22 genotypes, using ISSR analysis, yielded 117 fragments, of which 64 were polymorphic. Number of amplified fragments with ISSR primers ranged from five to ten and varied in size from 180 to 1,900 bp. Percentage polymorphism ranged from 0 to 87.5. The 15 RAPD primers produced 144 bands across 22 genotypes, of which 84 were polymorphic. The number of amplified bands varied from five to 13, with size range from 180 to 2,400 bp. Percentage polymorphism ranged from 0 to 100, with an average of 58.3 across. RAPD markers were relatively more efficient than the ISSR assay. The mental test between two Jaccard’s similarity matrices gave r ≥ 0.90, showing very good fit correlation in between ISSR- and RAPD-based similarities. Clustering of isolates remained more or less the same in RAPD and combined data of RAPD and ISSR. The similarity coefficient ranged from 0.734 to 0.939, 0.563 to 0.946, and 0.648 to 0.920 with ISSR, RAPD, and combined dendrogram, respectively.




Similar content being viewed by others
References
Arnau G, Lallemand J, Bourgoin M (2003) Fast and reliable strawberry cultivar identification using inter simple sequence repeat (ISSR) amplification. Euphytica 129:69–79
Bachmann K (1997) Nuclear DNA markers in plant biosystematics research. Opera Bot 132:137–148
Chen JM, Gituru WR, Wang YH, Wang QF (2006) The extent of clonality and genetic diversity in the rare Caldesia grandis (Alismataceae): comparative results for RAPD and ISSR markers. Aquat Bot 84:301–307
Chowdhury MA, Vandenberg B, Warkentin T (2002) Cultivar identification and genetic relationship among selected breeding lines and cultivars in chickpea (Cicer arietinum L.). Euphytica 127:317–325
Dellaport SL, Wood J, Hicks JB (1983) A plant DNA mini-preparation: version II. Plant Mol Biol Rep 1:19–21
Dwivedi SL, Gurtu S, Chandra S, Yuejin W, Nigam SN (2001) Assessment of genetic diversity among selected groundnut germplasm I: RAPD analysis. Plant Breed 120:345–349
Esselman EJ, Li JQ, Crawford D, Winduss JL, Wolfe AD (1999) Clonal diversity in the rare Calamagrostis porteri ssp. insperata (Poaceae): comparative results for allozymes and random amplified polymorphic DNA (RAPD) and inter-simple sequence repeat (ISSR) markers. Mol Ecol 8:443–451
Goulaõ L, Oliveira CM (2001) Molecular characterization of cultivars of apple (Malus domestica Borkh.) using microsatellite (SSR and ISSR) markers. Euphytica 122:81–89
Jaccard P (1908) Nouvelles recherches sur la distribution florale. Bull Soc Vaud Sci Nat 44:223–270
Joshi SP, Gupta VS, Aggarwal RK, Ranjekar PK (2000) Genetic diversity and phylogenetic relationship as revealed by inter simple sequence repeat (ISSR) polymorphism in the genus Oryza. Theor Appl Genet 100:1311–1320
Kumar P, Singh K, Vikal Y, Randhawa LS, Chahal GS (2003) Genetic diversity studies of elite cotton germplasm lines using RAPD markers and morphological characteristics. Indian J Genet 63(1):5–10
Landergott U, Holderegger R, Kozlowski G, Schneller JJ (2001) Historical bottlenecks decrease genetic diversity in natural populations of Dryopteris cristata. Heredity 87:344–355
Li A, Ge S (2001) Genetic variation and clonal diversity of Psammochloa villosa (Poaceae) detected by ISSR markers. Ann Bot 87:585–590
Mantel N (1967) The detection of disease clustering and generalized regression approach. Cancer Res 27:209–220
Martins M, Tenreiro R, Oliveira MM (2003) Genetic relatedness of Portuguese almond cultivars assessed by RAPD and ISSR markers. Plant Cell Reports 22:71–78
Parsons BJ, Newbury HJ, Jackson MT, Ford-Lloyd BV (1997) Contrasting genetic diversity relationships are revealed in rice (Oryza sativa L.) using different marker types. Mol Breed 3:115–125
Penner GA (1996) RAPD analysis of plant genomes. In: Jauhar PP (ed) Methods of genome analysis in plants. CRC, Boca Raton, pp 251–268
Raina SN, Rani V, Kojima T, Ogihara Y, Singh KP, Devarumath RM (2001) RAPD and ISSR fingerprints as useful genetic markers for analysis of genetic diversity, varietals identification, and phylogenetic relationships in peanut (Arachis hypogaea) cultivars and wild species. Genome 44:763–772
Rajaseger G, Tan HTW, Turner IM, Kumar PP (1997) Analysis of genetic diversity among Ixora cultivars (Rubiaceae) using random amplified polymorphic DNA. Ann Bot 80:355–361
Rohlf FJ (1993) NT-SYS-pc: numerical taxonomy and multivariate analysis system, version 2.11 W. Exteer Software, Setauket
Soller M, Beckmann JS (1983) Genetic polymorphism in varietal identification and genetic improvement. Theor Appl Genet 67:25–33
Souframanien J, Gopalakrishna T (2004) A comparative analysis of genetic diversity in black gram genotypes using RAPD and ISSR markers. Theor Appl Genet 109:1687–1693
Williams JGK, Kubelic AR, Livak KJ, Rafalski JA, Tingey SV (1990) DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res 18:6531–6535
Yap IV, Nelson RJ (1996) Winboot: a program for performing bootstrap analysis of binary data to determine the confidence of UPGMA-based dendrograms. IRRI, Manila
Yee E, Kidwell KK, Sills GR, Lumpkin TA (1999) Diversity among selected Vigna angularis (Azuki) accessions on the basis of RAPD and AFLP markers. Crop Sci 39:268–275
Zeitkiewicz E, Rafalski A, Labuda D (1994) Genome fingerprinting by simple sequence repeat (SSR)-anchored polymerase chain reaction amplification. Genomic 20:176–183
Acknowledgements
The authors are thankful to Dr. Ombir Singh, Division of Silviculture, Forest Research Institute, Dehradun, for providing the seed sources of different germplasms and to the Department of Biotechnology, Government of India, for providing financial support for conducting the investigation and fellowship to the first author.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Arif, M., Zaidi, N.W., Singh, Y.P. et al. A Comparative Analysis of ISSR and RAPD Markers for Study of Genetic Diversity in Shisham (Dalbergia sissoo). Plant Mol Biol Rep 27, 488–495 (2009). https://doi.org/10.1007/s11105-009-0097-0
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-009-0097-0