Abstract
Aims
Differential effects of the seed exudates of susceptible and resistant soybean cultivars Sloan and Williams 82 on the zoospores of soilborne pathogenic Phytophthora sojae are not clear. This study aims to elucidate the role of the seed exudates in the host resistance of soybean against P. sojae during prior to infection.
Methods
The response behaviors of P. sojae zoospores to the seed exudates were performed in an assay chamber and concave slide, the proteomes of P. sojae zoospores treated with the seed exudates were analyzed by TMT method. The expression of genes encoding key proteins was quantified by qRT-PCR.
Results
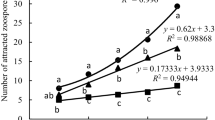
The zoospores of P. sojae exhibited significant chemotaxis to the both seed exudates and a stronger attracted was found to the seed exudates of Sloan. The seed exudates of Sloan also had a stonger promotion than that of Williams 82 on encystment of zoospores. During the responding process, the key proteins in zoospores related to chemotaxis were upregulated, the upregulation fold of these proteins in responding to Sloan was higher than that of Williams 82. Proteins differentially expressed promote the encystment of zoospores. The changes in expression of these proteins was several fold higher in the zoosprores exposed to the seed exudates of Sloan than that of Williams 82.
Conclusions
Evidence of differential changes in protein expression in P. sojae zoospores exposed to the seed exudates of both soybean supports the differential developmental behaviors such as chemotaxis and encystment of the zoospores in responding to the two seed exudates.






Similar content being viewed by others
Abbreviations
- MAPK:
-
Mitogen-activated protein kinase
- qRT-PCR:
-
quantitative Real-time polymerase chain reaction
- UPLC:
-
Ultra performance liquid chromatography
- LC-MS/MS:
-
Liquid chromatography-mass spectrometry/mass spectrometry
- Gα:
-
G protein α subunit
- GPCR:
-
G protein-coupled receptor
- TCA:
-
Trichloroacetic acid
- TEAB:
-
Tetraethylammonium bromide
- NSI:
-
Nanoelectrospray ionization
- TMT:
-
Tandem mass tag
- HPLC:
-
High performance liquid chromatography
- NCE:
-
Normalized collision energy
- AGC:
-
Automatic gain control
- Gβ :
-
G protein β subunit
- GO:
-
Gene Ontology
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- cv.:
-
Cultivar
- PI3K:
-
Phosphatidylinositol 3-kinase
- PI4K:
-
Phosphatidylinositol 4-kinase
- PKC:
-
Protein kinase C
References
Bahrenfus JB, Fehr WR (1980) Registration of Sloan soybean. Crop Sci 20(3):415–416
Barz W, Mackenbrock U (1994) Constitutive and elicitation induced metabolism of isoflavones and pterocarpans in chickpea (Cicer arietinum) cell suspension cultures. Plant Cell Tiss Org 38:199–211. https://doi.org/10.1007/bf00033878
Bernard RL, Cremeens CR (1988) Registration of “Williams 82” soybean. Crop Sci 28(6):1027–1028
Canaday HC (1979) The effects of nitrogen fertilization on Phytophthora rot of soybeans and on its causal pathogen, Phytophthora megasperma var. sojae. Thesis of Ohio State University, the U.S
Connolly MS, Williams N, Heckman CA, Morris PF (1999) Soybean isoflavones trigger a calcium influx in Phytophthora sojae. Fungal Genet Biol 28:6–11. https://doi.org/10.1006/fgbi.1999.1148
Dong S, Yu D, Cui L, Qutob D, Jones J, Kale S, Tyler BM, Wang Y, Gijzen M (2011) Sequence variants of the Phytophthora sojae RXLR effector avr3a/5 are differentially recognized by Rps3a and Rps5 in soybean. PloS One 6(7):e20172
Gao J, Cao M, Ye W, Li H, Kong L, Zheng X, Wang Y (2015) PsMPK7, a stress-associated mitogen-activated protein kinase (MAPK) in Phytophthora sojae, is required for stress tolerance, reactive oxygenated species detoxification, cyst germination, sexual reproduction and infection of soybean. Mol Plant Pathol 16:61–70. https://doi.org/10.1111/mpp.12163
Gong X, Jiang Y (2003) The structural basis of biological function of Mitogen-activated protein kinases. Chin J Biochem Mol Biol 19:5–11. https://doi.org/10.13865/j.cnki.cjbmb.2003.01.002 (in Chinese with English abstract)
Haudenshield JS, Song JY, Hartman GL (2017) A novel, multiplexed, probe-based quantitative PCR assay for the soybean root- and stem-rot pathogen, Phytophthora sojae, utilizes its transposable element. Plos One 12:e0176567. https://doi.org/10.1371/journal.pone.0176567
Hosseini S, Heyman F, Olsson U, Borberg A, Funck D, Karlsson M (2014) Zoospores chemotaxis of closely related legume-root infecing Phytophthora species towards host isoflavones. Plant Pathol 63:708–714. https://doi.org/10.1111/ppa.12137
Hua C (2008) Function analysis of G protein and calcium signal transduction pathways in Phytophthora sojae. Doctor degree dissertation of Nanjing Agricultural University, China (in Chinese with English abstract)
Hua C, Wang Y, Zheng X, Dou D, Zhang Z, Govers F, Wang Y (2008) A Phytophthora sojae G-protein alpha subunit is involved in chemotaxis to soybean isoflavones. Eukaryot Cell 7:2133–2140. https://doi.org/10.1128/EC.00286-08
Jun Y, Waseem R, Qirong S (2018) Root exudates dominate the colonization of pathogen and plant growth-promoting rhizobacteria. Soil Bio 52(6):167–180. https://doi.org/10.1007/978-3-319-75910-46
Kaitany R, Melakeberhan H, Bird GW, Safir G (2000) Association of Phytophthora sojae with Heterodera glycines and nutrient stressed soybeans. Nematropica 30(2):193–200
Khafizov K, Lattanzi G, Carloni P (2009) G protein inactive and active forms investigated by simulation methods. Proteins 75:919–930. https://doi.org/10.1002/prot.22303
Kittle DR, Gray LE (1979) The influence of soil temperature, moisture, porosity, and bulk density on the pathogenicity of Phytophthora megasperma var. sojae. Plant Dis 63:231–234
Li A, Wang Y, Tao K, Dong S, Huang Q, Dai T, Zheng X, Wang Y (2010) PsSAK1, a stress activated MAP kinase of Phytophthora sojae, is required for zoospore viability and infection of soybean. Mol Plant Microbe In 23:1022–1031. https://doi.org/10.1094/MPMI-23-8-1022
Li L, Yang M, Cui R, Song C, Wen J (2012) Transformation and expression of enhanced green fluorescent protein gene in Phytophthora sojae (in Chinese with English summary). Acta Phytophylacica Sinica 39:59–60
Lin C, Aronson JM (1970) Chitin and cellulose in the cell walls of the Oomycete Apodachlya sp. Arch Mikrobiol 72:111–114. https://doi.org/10.1007/BF00409517
Liu J, Baloucoune GA, Chun L (2012) Palmitoylation modification regulates G-protein-coupled receptors effects. Chin J Biochem Mol Biol 28:99–107 (in Chinese with English abstract)
Morris PF, Ward EWB (1992) Chemoattraction of zoospores of the soybean pathogen, Phytophthora sojae, by isoflavones. Physiol Mol Plant Pathol 40:17–22. https://doi.org/10.1016/0885-5765(92)90067-6
Schmitthenner AF (1985) Problem and progress in control of Phytophthora root rot of soybean. Plant Dis 69:362–368. https://doi.org/10.1094/PD-69-362
Shen G (2005) Differentially expressed genes during early infection of Phytophthora sojae with soybean plant and funcional analysis of some genes. Doctor degree dissertation of Nanjing Agricultural University, China (in Chinese with English abstract)
Suo B, Cui R, Tian M, Wen J (2015) Effects of soil environment on activation of oospores of Phytophthora sojae. J Plant Protect 42(3):370–375. https://doi.org/10.13802/j.cnki.zwbhxb.2015.03.013. (in Chinese with English abstract)
Suo B, Chen Q, Wu W, Wu D, Tian M, Jie Y, Zhang B, Wen J (2016) Chemotactic responses of Phytophthora sojae zoospores to amino acids and sugars in root exudates. J Gen Plant Pathol 82:142–148
Tyler BM (2007) Phytophthora sojae: root rot pathogen of soybean and model oomycete. Mol Plant Pathol 8:1–8. https://doi.org/10.1111/j.1364-3703.2006.00373.x
Wagner R, Norris S, Chatterjee P, Morris PF, Wildschutte H (2018) Aquatic Pseudomonads inhibit oomycete plant pathogens of Glycine max. Front Microbiol 9:1007
Wang Y, Zheng X (2007) Study on the mutual recognition and pathogenic mechanism between Phytophthora sojae and its host. In: Abstracts of the first National Symposium on biological invasion. Fuzhou, China, pp5 (in Chinese)
Wang Y, Li A, Wang X, Zhang X, Zhao W, Dou D, Zheng X, Wang Y (2010) GPR11, a putative seven-transmembrane G protein-coupled receptor, controls zoospore development and virulence of Phytophthora sojae. Eukaryot Cell 9:242–250. https://doi.org/10.1128/ec.00265-09
Wu W, Chen Q, Chen Y, Wu D, Gao X, Zhao Y, Xu Y, Zhang Z, Song G, Wen J (2018) Response of Phytophthora sojae zoospores to the rootlet and root exudates of host soybean and non-host common bean and host selectivity. Microbiol China 45:1491–1499. https://doi.org/10.13344/j.microbiol.china.170990 (in Chinese with English abstract)
Yang Z, Yang Q, Chen C (2011) The research progress in the chemokine SDF-1 and the chemokine receptor CXCR4. J Mol Diagn Ther 3:58–60. https://doi.org/10.3969/j.issn.1674-6929.2011.01.015 (in Chinese with English abstract)
Yang X, Zhao W, Hua C (2013) Chemotaxis and oospore formation in Phytophthora sojae are controlled by G-protein-coupled receptors with a phosphatidylinositol phosphate kinase domain. Mol Microbiol 88:382–394. https://doi.org/10.1111/mmi.12191
Yannis R, Michael WD (2006) Zoospores encystment and pathogenicity of Phytophthora and Pythium species on plant roots. Microbiol Res 161(1):1–8. https://doi.org/10.1016/j.micres.2005.04.003
Ye W, Li A, Wang X, Wang Y (2016) Identification and transcriptional analysis of the MAPK genes in Phytophthora sojae. Acta Phytopathol Sin 46:338–346. https://doi.org/10.13926/j.cnki.apps.2016.03.007 (in Chinese with English abstract)
Zhang X (2015) Molecular mechanisms of Phytophthora sojae PsGPA1 in regulation of zoospore chemotaxis to soybean. Doctor degree dissertation of Nanjing Agricultural University, China (in Chinese with English abstract)
Zhang Z, Xu Y, Song G, Gao X, Zhao Y, Jia M, Chen Y, Suo B, Chen Q, Wu D, Wu W, Wen J (2019) Phytophthora sojae zoospores differ in chemotaxis to the root and root exudates of host soybean and nonhost common bean. J Gen Plant Pathol 85:201–210. https://doi.org/10.1007/s10327-019-00839-9
Zuo Y, Kang Z, Huang L, Han Q (2005) Cytology on infection process of soybean hypocotyls by Phytophthora sojae. Acta Phytopathol Sin 35:235–241. https://doi.org/10.3321/j.issn:0412-0914.2005.03.008 (in Chinese with English abstract)
Acknowledgements
Thanks for the technical support and bioinformatics analysis of PTB BioLabs.
Funding
This research was supported by the National Natural Science Foundation of China (Grant No. 31670444; 31370449) to Jingzhi Wen.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were mainly performed by Zhuoqun Zhang, Haixu Liu, Xiangqi Bi, Han Yu, Ying Xu, Yufei Chen, and Zhiyue Yang. The first draft of the manuscript was written by Zhuoqun Zhang with help from Jingzhi Wen, and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Responsible Editor: Matthew G. Bakker
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Zhang, Z., Liu, H., Bi, X. et al. Differential response of Phytophthora sojae zoospores to soybean seed exudates provides evidence of seed exudates participate in host resistance. Plant Soil 452, 601–614 (2020). https://doi.org/10.1007/s11104-020-04607-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-020-04607-z