Abstract
Background and aims
Production of indole-3-acetic acid (IAA) by Azospirillum brasilense is one of the most important mechanisms underlying the beneficial effects observed in plants after inoculation with this bacterium. This study determined the contribution of the hisC1 gene, which encodes aromatic amino acid aminotransferase-1 (AAT1), to IAA production, and analyzed its expression in the free-living state and in association with the roots of wheat.
Methods
We determined production of IAA and AAT activity in the mutant hisC::gusA-sm R. To study the expression of hisC1, a chromosomal gene fusion was analyzed by following β-glucuronidase (GUS) activity in vitro, in the presence of root exudates, and in association with roots.
Results
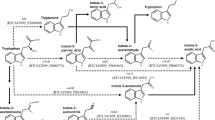
IAA production in the hisC mutant was not reduced significantly compared to the activity of the wild-type strain. AAT1 activity was reduced by 50% when tyrosine was used as the amino acid donor, whereas there was a 30% reduction when tryptophan was used, compared to the activity of the wild-type strain. Expression of the fusion protein was up-regulated in both logarithmic and stationary phases by several compounds, including IAA, tryptophan, tyrosine, and phenyl acetic acid. We observed the expression of hisC1 in bacteria associated with wheat roots. Root exudates of wheat and maize were able to stimulate hisC1 expression.
Conclusions
The expression data indicate that hisC1 is under a positive feedback control in the presence of root exudates and on plants, suggesting that AAT1 activity plays a role in Azospirillum–plant interactions.





Similar content being viewed by others
Abbreviations
- IAA:
-
Indole-3-acetic acid
- AAT1:
-
Aromatic amino acid aminotransferase-1
- IPyA:
-
Indole-3-pyruvic acid
- IAdhl:
-
Indole-3-acetaldehyde
- Trp:
-
Tryptophan
- Tyr:
-
Tyrosine
- Phe:
-
Phenylalanine
- pHPP:
-
p-Hydroxyphenylpyruvate
- PAA:
-
Phenyl acetic acid
- HPLC:
-
High-performance liquid chromatography
References
Alexeyev MF, Shokolenko IN, Croughan TP (1995) Improved antibiotic-resistance gene cassettes and omega elements for Escherichia coli vector construction and in vitro deletion/insertion mutagenesis. Gene 160:63–67
Arsène F, Katupitiya S, Kennedy IR, Elmerich C (1994) Use of lacZ fusions to study the expression of nif genes of Azospirillum brasilense in association with plants. Mol Plant Microbe Interact 7:748–757
Baca EB, Soto-Urzúa L, Xochihua YG, Cuervo A (1994) Characterization of aromatic amino acid aminotransferases and production of indolacetic acid in Azospirillum strains. Soil Biol Biochem 26:57–63
Bashan Y, de-Bashan L (2010) How the plant growth-promoting bacterium Azospirillum promotes plant growth—a critical assessment. Adv Agron 108:77–136
Bashan Y, Harrison SK, Whitmoyer RE (1990) Enhanced growth of wheat and soybean plants inoculated with Azospirillum brasilense is not necessarily due to general enhancement of mineral uptake. Appl Environ Microbiol 56:769–775
Bashan Y, Holguin G, de-Bashan LE (2004) Azospirillum–plant relationships: physiological, molecular, agricultural, and environmental advances (1997–2003). Can J Microbiol 50:521–577
Carreño López R, Campos Reales N, Elmerich C, Baca BE (2000) Physiological evidence for differently regulated tryptophan-dependent pathways for indole-3-acetic acid synthesis in Azospirillum brasilense Sp7. Mol Gen Genet 264:521–530
Cohen AC, Bottini R, Piccoli PN (2008) Azospirillum brasilense Sp 245 produces ABA in chemically-defined culture medium and increases ABA content in arabidopsis plants. Plant Growth Regul 54:97–103
Costacurta A, Keijers V, Vanderleyden J (1994) Molecular cloning and sequence analysis of an Azospirillum brasilense indole-3-pyruvate decarboxylase gene. Mol Gen Genet 243:463–472
Creus CM, Graziano M, Casanovas EM, Pereyra MA, Simontacchi M, Puntarulo S, Barassi CA, Lamattina L (2005) Nitric oxide is involved in the Azospirillum brasilense-induced lateral root formation in tomato. Planta 221:297–303
Crozier A, Arruda P, Jasmin JM, Monterio AM,Sandberg G (1988) Analysis of indole-3 acetic acidand related indoles in culture medium and Azospirillumbrasilense. Appl Environ Microbiol 54:2833–2837
Diamondstone TI (1966) Assay of tyrosine transaminase activity by conversion of p-hydroxyphenylpyruvate to hydroxybenzaldehyde. Anal Biochem 16:395–401
Dobbelaere S, Croonenborghs A, Thys A, Ptacek D, Vanderleyden J, Dutto P et al (2001) Responses of agronomically important crops to inoculation with Azospirillum. Aust J Plant Physiol 28:871–879
Frankenberger WT, Arshad M (1995) Phytohormones in soil: microbial production and function. Dekker, New York
Ge SM, Li T, Chen SF (2009) Expression and functional analysis of aminotransferase involved in indole-3-acetic acid biosynthesis in Azospirillum brasilense Y62. Biochem (Moscow) 74:81–84
González-López J, Rodelas B, Pozo C, Salmerón-López V, Martínez-Toledo MV, Salmerón V (2005) Liberation of amino acids by heterotrophic nitrogen fixing bacteria. Amino Acids 28:363–367
Granner DK, Tomkins GM (1970) Tyrosine aminotransferase (rat liver). Methods Enzymol 17:633–637
Hoffland E (1992) Quantitative evaluation of the role of organic acid exudation in the mobilization of rock phosphate by rape. Plant Soil 140:279–289
Jensen RA, Gu W (1996) Evolutionary recruitment of biochemically specialized subdivisions of Family I within the protein superfamily of aminotransferases. J Bacteriol 178:2161–2171
Kamilova F, Kravchenko LV, Shaposhnikov AI, Azarova T, Makarova N, Lugtenberg B (2006) Organic acids, sugars and L-tryptophane in exudates and vegetables growing on stonewool and their effects on activities of rhozosphere bacteria. Mol Plant Microbe Interact 19:250–256
Kaneko T et al (2010) Complete genomic structure of the cultivated rice endophyte Azospirillum sp B510. DNA Res 17:37–50. doi:10.1093/dnares/dsp026
Koga J, Syono K, Ichikawa T, Adachi T (1994) Involvement of l-tryptophan in indole-3-acetic acid biosynthesis in Enterobacter cloacae. Biochem Biophys Acta 1209:241–247
Lynch JM, Whipps JM (1990) Substrate flow in the rhizosphere. Plant Soil 129:1–10
Malhotra M, Srivastava S (2008) organization of the ipdC region regulates IAA levels in different Azospirillum brasilense strains: molecular and functional analysis of ipdC in strain SM. Environ Microbiol 10:1365–1373
Martínez-Morales LJ, Soto-Urzúa L, Baca BE, Sánchez-Ahédo JA (2003) Indole-3-butyric acid (IBA) production in culture medium by wild-type Azospirillum brasilense. FEMS Microbiol Lett 228:167–173
Matilla MA, Espinosa-Urgel M, Rodríguez-Herva JJ, Ramos JL, Ramos-González MI (2007) Genomic analysis reveals the major driving forces of bacterial life in the rhizosphere. Genome Biol 8:R1791–13
Miller JH (1972) Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NewYork
Murcia R, Rodelas B, Salmerón V, Pozo C, González-López J (1997) Effect of simazine on the production of lisine and methionine by Azotobacter chroococcum and Azotobacter vinelandii. Amino Acids 12:249–255
Okon Y, Labandera-González CA (1994) Agronomic applicationsof Azospirillum: an evaluation of 20 years worldwide fieldinoculation. Soil Biol Biochem 26:1591–1601
Van Overbeek LS, Van Elsas JD (1995) Root exudate-induced promoter activity in Pseudomonas fluorescensmutants in the wheat rhizosphere. Appl Environ Microbiol 61:889–898
Pedraza RO, Ramírez Mata A, Xiqui Vázquez ML, Baca BE (2004) Aromatic amino acid aminotransferase activity and índole-3-acetic acid production by associate nitrogen-fixing bacteria. FEMS Microbiol Lett 233:15–21
Phillips DA, Fox TC, King MD, Bhuvaneswari TV, Teuber LR (2004) Microbial products trigger amino acid exudation from plant roots. Plant Physiol 136:2887–2894
Piccoli P, Masciarelli O, Bottini R(1996) Metabolism of 17,17-[2H2]-gibberellin A4, A9 and A20 by Azospirillum lipoferum in chemically defined culture medium. Symbiosis 21:263–274
Rothballer M, Schmid M, Fekete A, Hartmann A (2005) Comparative in situ analysis of ipdC-gfpmut3 promoter fusions of Azospirillum brasilense strains Sp7 and Sp245. Environ Microbiol 7:1839–1846
Ruckdäschel E, Lewis-Kittell B, Helinski DR, Klingmüller W (1988) Aromatic amino acid aminotransferase of Azospirillum lipoferum and their possible involvement in IAA biosynthesis. In: Klingmüller W (ed) Azospirillum IV, genetics, physiology, ecology. Springer, The Netherlands, pp 49–53
Ryu RJ, Patten CL (2008) Aromatic amino acid dependent expression of índole-3-pyruvate decarboxylase is regulated by TyrR in Enterobacter cloacae UW5. J Bacteriol 190:7200–7208
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York
Sergeeva E, Hirkala DLM, Nelson LM (2007) Production of indole-3-acetic acid, aromatic amino aminotransferase activities and plant growth promotion by Pantoea agglomerans rhizosphere isolates. Plant Soil 297:1–13
Simon R, Priefer U, Pühler A (1983) A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Biotechnology 1:784–790
Somers E, Ptacek D, Gysegom P, Srinivasan M, Vanderleyden J (2005) Azospirillum brasilense produces the auxin-like phenylacetic acid by using the key enzyme for indol-3-acetic acid biosynthesis. Appl Environ Microbiol 71:1803–1810
Soto-Urzúa L, Xochihua-Corona YG, Flores-Encarnación M, Baca BE (1996) Purification and properties of aromatic amino acid aminotransferase from Azospirillum brasilense UAP14 strain. Can J Microbiol 42:294–298
Spaepen S, Vanderleyden J, Remans R (2007a) Indole-3-acetic acid in microbial and microorganism-plant signaling. FEMS Microbiol Rev 31:425–448
Spaepen S, Versées W, Gocke D, Pohl M, Steyaert J, Vanderleyden J (2007b) Characterization of phenylpyruvate decarboxylase, involved in auxin production of Azospirillum brasilense. J Bacteriol 189:7626–7633
Spaepen S, Dobbelaere S, Croonenborghs A, Vanderleyden J (2008) Effects of Azospirillum brasilense of indol-e-3acetic-acid production on inoculated wheat plants. Plant Soil 312:15–23
Tao Y et al (2008) Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133:164–176
Tien TM, Gaskins MH, Hubbell DH (1979) Plant growth substances23 produced by Azospirillum brasilense and their effect on the growth of pearl millet24 (Pennisetum americanum L.). Appl Environ Microbiol 37:1016–1024
Vande Broek A, Michiels J, Van Gool A, Vanderleyden J (1993) Spatial-temporal colonization patterns of Azospirillum brasilense on the wheat root surface and expression of bacterial nifH gene during association. Mol Plant Microbe Interact 6:592–600
Vande Broek A, Lambrecht M, Eggermont K, Vanderleyden J (1999) Auxins up-regulate expression of the indole-3-pyruvate decarboxylase gene in Azospirillum brasilense. J Bacteriol 181:1338–1342
Vílchez S, Molina L, Ramos C, Ramos JL (2000) Proline catabolism by Pseudomonas putida: cloning, characterization, and expression of the put genes in the presence of root exudates. J Bacteriol 182:91–99
Yang S, Zhang Q, Guo J, Charkowski AM, Glick BR, Ibekwe AM, Cooksey DA, Yang CH (2007) Global effect of indole-3-acetic acid biosynthesis on multiple factors of Erwinia chrysanthemi 3937. Appl Environ Microbiol 73:1079–1088
Zeman AMM, Tchan YT, Elmerich C, Kennedy IR (1992) Nitrogenase activity in wheat seedlings bearing para-nodules induced by 2,4-dichlorophenoxyacetic acid (2,4-D) and inoculated with Azospirillum. Res Microbiol 143:847–855
Acknowledgments
This work was supported by a Consejo Nacional de Ciencia y Tecnología (CONACyT) Grant 49227-Z, together with Vicerrectoría de investigación y estudios de posgrado-Secretaría de educación pública (VIEP-SEP), Grant NAT-IC-1. J.G.C. was the recipient of a scholarship from CONACyT.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Euan K. James.
Rights and permissions
About this article
Cite this article
Castro-Guerrero, J., Romero, A., Aguilar, J.J. et al. The hisC1 gene, encoding aromatic amino acid aminotransferase-1 in Azospirillum brasilense Sp7, expressed in wheat. Plant Soil 356, 139–150 (2012). https://doi.org/10.1007/s11104-011-1009-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-011-1009-2