Abstract
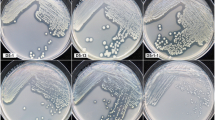
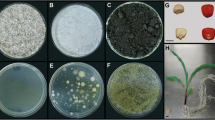
Sixty-six atrazine-degrading bacterial communities utilizing atrazine as sole N source and citrate as principal C source were isolated from unplanted and maize planted soils treated with atrazine. Ribosomal intergenic spacer analysis (RISA) fingerprints revealed that the genetic structure of atrazine-degrading bacterial communities was modified in the maize rhizosphere. To assess the underlying microbial diversity, 16S rDNA sequences amplified from each bacterial community were cloned. Libraries containing 660 16S rDNA clones were screened by restriction fragment length polymorphism (RFLP) analysis. In all, 63 clone families were identified. Rarefaction curves did not reach a clear saturation, indicating that the analysis of a greater number of clones would have revealed further diversity. Recovered 16S rDNA sequences were related to Actinobacteria, Bacteroidetes and Proteobacteria. The four dominant RFLP families were highly similar to Variovorax paradoxus, Burkholderia cepacia, Arthrobacter sp. and Bosea sp. The composition of most of the atrazine-degrading bacterial communities consisted of 2–7 different bacterial species. Various atrazine-degrading gene compositions were observed, two of these atzABCDEF, trzND and atzBCDEF, trzN being largely dominant. The first was more frequently detected in bacterial communities isolated from the maize rhizosphere whereas the second was more frequently detected in communities isolated from bulk soil. Monitoring of atrazine-degrading activity showed that 76% of the bacterial communities degraded up to 80% of the initially added atrazine within 15 days of culture. Altogether our results indicate that the maize rhizosphere has an impact on the genetic structure, the diversity and atrazine-degrading gene composition of the atrazine-degrading communities.
Similar content being viewed by others
References
E Barriuso S Houot (1996) ArticleTitleRapid mineralization of s-triazine ring of atrazine in soils in relation to soil management Soil Biol. Biochem. 28 1341–1348 Occurrence Handle1:CAS:528:DyaK2sXos1SgsQ%3D%3D Occurrence Handle10.1016/S0038-0717(96)00144-7
J Borneman E Triplett (1997) ArticleTitleMolecular microbial diversity in soils from Eastern Amazonia: evidence for unusual micro-organisms and microbial population shifts associated with deforestation Appl. Environ. Microb. 63 2647–2653 Occurrence Handle1:CAS:528:DyaK2sXktlektLg%3D
K L Boundy-Mills M L Souza Particlede R T Mandelbaum L P Wackett M J Sadowsky (1997) ArticleTitleThe atzB gene of Pseudomonas sp. strain ADP encodes the second enzyme of a novel atrazine degradation pathway Appl. Environ. Microb. 63 916–923 Occurrence Handle1:CAS:528:DyaK2sXhsV2mtbk%3D
C Bouquard J Ouazzni J C Promé Y Michel-Briand P Plésiat (1997) ArticleTitleDechlorination of atrazine by a Rhizobium sp. isolate Appl. Environ. Microb. 63 862–866 Occurrence Handle1:CAS:528:DyaK2sXhsV2jtLo%3D
B T Bowman (1989) ArticleTitleMobility and persistence of the herbicides atrazine, metolachlor and terbuthylazine in plainfield sand determined using field lysimeters Environ. Toxicol. Chem. 8 485–491 Occurrence Handle1:CAS:528:DyaL1MXksFagtb8%3D
B Cai Y Han B Liu Y Ren S Jiang (2003) ArticleTitleIsolation and characterization of an atrazine-degrading bacterium from industrial wastewater in China Lett. Appl. Microbiol. 36 272–276 Occurrence Handle12680937 Occurrence Handle1:CAS:528:DC%2BD3sXktVWjsbo%3D Occurrence Handle10.1046/j.1472-765X.2003.01307.x
D Cheneby L Philipot A Hartmann C Henault J C Germon (2000) ArticleTitle16S rDNA analysis for characterization of denitrifying bacteria isolated from three agricultural soils FEMS Microbiol. Ecol. 34 121–128 Occurrence Handle11102689 Occurrence Handle1:CAS:528:DC%2BD3cXotlCqsbo%3D
D Cheneby S Hallet M Mondon F Martin-Laurent J C Germon L Philippot (2003) ArticleTitleGenetic characterisation of the nitrate reducing community based on narG nucleotide sequences analysis Microbial Ecol. 46 113–121 Occurrence Handle1:CAS:528:DC%2BD3sXnt1Git7w%3D Occurrence Handle10.1007/s00248-002-2042-8
G Cheng N Shapir M J Sadowsky L P Wackett (2005) ArticleTitleAllophanate hydrolase, not urease, functions in bacterial cyanuric acid metabolism Appl. Environ. Microbiol. 71 4437–4445 Occurrence Handle16085834 Occurrence Handle1:CAS:528:DC%2BD2MXoslGitbs%3D Occurrence Handle10.1128/AEM.71.8.4437-4445.2005
E Curl B Truelove (1986) The rhizosphere Springer Berlin
M L Souza Particlede L P Wackett K L Boundy Mills R T Mandelbaum M J Sadowsky (1995) ArticleTitleCloning, characterization, and expression of a gene region from Pseudomonas sp. strain ADP involved in the dechlorination of atrazine Appl. Environ. Microb. 61 3373–3378
M L Souza Particlede J Seffernick B Martinez M J Sadowsky L P Wackett (1998a) ArticleTitleThe atrazine catabolism genes atzABC are widespread and highly conserved J. Bacteriol. 180 1951–1954
M L Souza Particlede L P Wackett M J Sadowsky (1998b) ArticleTitleThe atzABC genes encoding atrazine catabolism are located on a self transmissible plasmid in Pseudomonas sp. strain ADP Appl. Environ. Microb. 64 2323–2326
W Dejonghe E Berteloot J Goris N Boon K Crul S Maertens M Höfte P Vos Particlede W Verstraete E M Top (2003) ArticleTitleSynergistic degradation of linuron by a bacterial consortium and isolation of a single linuron-degrading Variovorax strain Appl. Environ. Microb. 69 1532–1541 Occurrence Handle1:CAS:528:DC%2BD3sXitlCls7s%3D Occurrence Handle10.1128/AEM.69.3.1532-1541.2003
M Devers G Soulas F Martin-Laurent (2004) ArticleTitleReal-time reverse transcription PCR analysis of expression of atrazine catabolism genes in two bacterial strains isolated from soil J. Microbiol. Meth. 56 3–15 Occurrence Handle1:CAS:528:DC%2BD2cXoslWn Occurrence Handle10.1016/j.mimet.2003.08.015
M Devers H Henry A Hartmann F Martin-Laurent (2005) ArticleTitleHorizontal gene transfer of atrazine-degrading genes (atz) from Agrobacterium tumefaciens St96–4 pADP1::Tn5 to bacteria of maize-cultivated soil Pest Manag. Sci. 61 870–880 Occurrence Handle16032656 Occurrence Handle1:CAS:528:DC%2BD2MXpvFSrurg%3D Occurrence Handle10.1002/ps.1098
L E Erickson K H Lee (1989) ArticleTitleDegradation of atrazine and related s-triazines Crit. Rev. Env. Contr. 12 1–14 Occurrence Handle10.1080/10643388909388356
F S Fadullon J S Karns A Torrents (1998) ArticleTitleDegradation of atrazine in soil by Streptomyces J. Environ. Health 33 37–49 Occurrence Handle1:STN:280:DyaK1c7lt1alug%3D%3D Occurrence Handle10.1080/03601239809373128
I Fruchey N Shapir M Sadowsky L P Wackett (2003) ArticleTitleOn the origins of cyanuric acid hydrolase: purification, substrates, and prevalence of AtzD from Pseudomonas sp. strain ADP Appl. Environ. Microb. 69 3653–3657 Occurrence Handle1:CAS:528:DC%2BD3sXks1Gksrg%3D Occurrence Handle10.1128/AEM.69.6.3653-3657.2003
E A Greene P H Beatty P M Fedorak (2000) ArticleTitleSulfolane degradation by mixed cultures and a bacterial isolate identified as a Variovorax sp Arch. Microbiol. 174 111–119 Occurrence Handle10985750 Occurrence Handle1:CAS:528:DC%2BD3cXlvVGisrk%3D Occurrence Handle10.1007/s002030000184
V Gurtler V A Stanisich (1996) ArticleTitleNew approaches to typing and identification of bacteria using the 16S–23S rDNA spacer region Microbiology 142 3–16 Occurrence Handle8581168 Occurrence Handle10.1099/13500872-142-1-3
K Kannan S Tannabe R J Williams R Tatsukawa (1994) ArticleTitlePersistent organochlorine residues in foodstuffs from Australia, Papua New Guinea and the Solomon Islands: contamination levels and dietary exposure Sci. Total Environ. 153 29–49 Occurrence Handle7939620 Occurrence Handle1:CAS:528:DyaK2cXlslyisbo%3D Occurrence Handle10.1016/0048-9697(94)90099-X
J S Karns (1999) ArticleTitleGene sequence and properties of an s-triazine ring-cleavage enzyme from Pseudomonas sp. strain NRRLB-12227 Appl. Environ. Microb. 65 3512–3517 Occurrence Handle1:CAS:528:DyaK1MXltVOlsbs%3D
T J Kim E Y Lee Y J Kim K S Cho H W Ryu (2003) ArticleTitleDegradation of polyaromatic hydrocarbons by Burkholderia cepacia 2A-12 World J. Microb. Biot. 19 411–417 Occurrence Handle1:CAS:528:DC%2BD3sXktFSmtbo%3D Occurrence Handle10.1023/A:1023998719787
G A Kowalchuck D S Buma W D Boer P G Klinkhamer J A Veen Particlevan (2002) ArticleTitleEffects of above-ground plant species composition and diversity on the diversity of soil-borne micro-organisms A. van Leeuw. J. Microb. 81 509–520 Occurrence Handle10.1023/A:1020565523615
C R Kuske L O Ticknor M E Miller J D Dunbar J A Davis S M Barns J Belnap (2002) ArticleTitleComparison of soil bacterial communities in rhizospheres of three plant species and the interspaces in an arid grassland Appl. Environ. Microb. 68 1854–1863 Occurrence Handle1:CAS:528:DC%2BD38XivFGlur4%3D Occurrence Handle10.1128/AEM.68.4.1854-1863.2002
H Baron ParticleLe (1982) Herbicide Resistance in Plants John Wiley & Sons New York
R T Mandelbaum D L Allan L P Wackett (1995) ArticleTitleIsolation and characterization of a Pseudomonas sp. that mineralizes the s-triazine herbicide atrazine Appl. Environ. Microb. 61 1451–1457 Occurrence Handle1:CAS:528:DyaK2MXksl2lsLk%3D
F Martin-Laurent P Franken S Gianinazzi (1995) ArticleTitleScreening of cDNA fragments generated by differential RNA display Anal. Biochem. 228 182–184 Occurrence Handle8572283 Occurrence Handle1:CAS:528:DyaK2MXmsFyrtLw%3D Occurrence Handle10.1006/abio.1995.1337
F Martin-Laurent L Philippot S Hallet R Chaussod J C Germon G Soulas G Catroux (2001) ArticleTitleDNA extraction from soils: old bias for new microbial diversity analysis methods Appl. Environ. Microb. 67 2354–2359 Occurrence Handle1:CAS:528:DC%2BD3MXjtlGmsLg%3D Occurrence Handle10.1128/AEM.67.5.2354-2359.2001
F Martin-Laurent S Piutti S Hallet I Wagschal L Philippot G Catroux G Soulas (2003) ArticleTitleQuantitative competitive PCR as a tool to monitor the atzC, 16S and 18S rDNA genes from soil Pest Manag. Sci. 59 1–10 Occurrence Handle10.1002/ps.630 Occurrence Handle1:CAS:528:DC%2BD3sXitlCntbk%3D
B Martinez J Tomkins L P Wackett R Wing M J Sadowsky (2001) ArticleTitleComplete nucleotide sequence and organization of the atrazine catabolic plasmid pADP-1 from Pseudomonas sp. strain ADP J. Bacteriol. 183 5684–5697 Occurrence Handle11544232 Occurrence Handle1:CAS:528:DC%2BD3MXmvF2msL0%3D Occurrence Handle10.1128/JB.183.19.5684-5697.2001
L L H Mc Cormick (1966) ArticleTitleMicrobiological decomposition of atrazine and diuron in soil Weeds 14 77–82 Occurrence Handle1:CAS:528:DyaF28XntV2htQ%3D%3D
W W Mulbry H Zhu S M Nour E Topp (2002) ArticleTitleThe triazine hydrolase gene trzN from Nocardioides sp. strain C190: cloning and construction of gene-specific primers FEMS Microbiol. Lett. 206 75–79 Occurrence Handle11786260 Occurrence Handle1:CAS:528:DC%2BD38XitFahsA%3D%3D
S R Muller M Berg M M Ulrich R P Schwarzenbach (1997) ArticleTitleAtrazine and its primary metabolites in Swiss lakes: input characteristics and long-term behaviour in water column Environ. Sci. Technol. 31 2104–2113 Occurrence Handle10.1021/es9609314
L Philippot S Piutti F Martin-Laurent S Hallet J Germon (2002) ArticleTitleMolecular analysis of the nitrate-reducing community from unplanted and maize-planted soils Appl. Environ. Microb. 68 6121–6128 Occurrence Handle1:CAS:528:DC%2BD38XptlartL4%3D Occurrence Handle10.1128/AEM.68.12.6121-6128.2002
F E Pick L P Van E Botha (1992) ArticleTitleAtrazine in ground and surface water in maize production areas of the Transvaal South Africa Chemosphere 25 335–341 Occurrence Handle1:CAS:528:DyaK38XlvFyjsLg%3D Occurrence Handle10.1016/0045-6535(92)90550-B
S Piutti S Hallet S Rousseaux L Philippot G Soulas F Martin-Laurent (2002a) ArticleTitleAccelerated biodegradation of atrazine in soil cultivated with maize Biol. Fert. Soils 36 434–441 Occurrence Handle1:CAS:528:DC%2BD38XptlGhs7c%3D Occurrence Handle10.1007/s00374-002-0545-6
S Piutti A L Marchand B Lagacherie F Martin-Laurent G Soulas (2002b) ArticleTitleEffect of cropping cycles and repeated herbicide applications on the degradation of diclofop-methyl, bentazone, diuron, isoproturon and pendimethalin in soil Pest Manag. Sci. 58 303–312 Occurrence Handle1:CAS:528:DC%2BD38XitFSgtLg%3D Occurrence Handle10.1002/ps.459
S Piutti E Semon D Landry A Hartmann S Dousset E Lichtfouse E Topp G Soulas F Martin-Laurent (2003) ArticleTitleIsolation and characterisation of Nocardioides sp. SP12, an atrazine degrading bacterial strain possessing the gene trzN from bulk and maize rhizosphere soil FEMS Microbiol. Lett. 221 111–117 Occurrence Handle12694918 Occurrence Handle1:CAS:528:DC%2BD3sXivFCru74%3D Occurrence Handle10.1016/S0378-1097(03)00168-X
E Robleto J Borneman E W Triplett (1998) ArticleTitleEffect of bacterial antibiotic production on rhizosphere microbial communities from a culture independent perspective Appl. Environ. Microb. 64 5020–5022 Occurrence Handle1:CAS:528:DyaK1cXnvF2htLc%3D
S Rousseaux A Hartmann G Soulas (2001) ArticleTitleIsolation and characterisation of new Gram-negative and Gram-positive atrazine degrading bacteria from different French Soils FEMS Microbiol. Ecol. 36 211–222 Occurrence Handle11451526 Occurrence Handle1:CAS:528:DC%2BD3MXltVeisL0%3D
S Rousseaux G Soulas A Hartmann (2002) ArticleTitlePlasmid localisation of atrazine-degrading genes in newly described Chelatobacter and Arthrobacter strains FEMS Microbiol. Ecol. 1371 1–7
M J Sadowsky Z Tong M L Souza Particlede L P Wackett (1998) ArticleTitleAtzC is a new member of the aminohydrolase protein superfamily and is homologous to other atrazine-metabolizing enzymes J. Bacteriol. 180 152–158 Occurrence Handle9422605 Occurrence Handle1:CAS:528:DyaK1cXotVWm
E Simpson (1949) ArticleTitleMeasurement of diversity Nature 163 688
A Schmalenberger C Tebbe (2003) ArticleTitleBacterial diversity in maize rhizosphere: conclusions on the use of genetic profiles based on PCR-amplified partial small sub-unit rRNA genes in ecological studies Mol. Ecol. 12 251–262 Occurrence Handle12492893 Occurrence Handle1:CAS:528:DC%2BD3sXht1Sjur0%3D Occurrence Handle10.1046/j.1365-294X.2003.01716.x
D R Shelton S Khader B M Pogell (1996) ArticleTitleMetabolism of twelve herbicides by Streptomyces Biodegradation 7 129–136 Occurrence Handle8882805 Occurrence Handle1:CAS:528:DyaK28XjtlSgtL0%3D Occurrence Handle10.1007/BF00114625
M S Shields M J Reagin R R Gerger R Campbell C Somerville (1995) ArticleTitleTOM, a new aromatic degradative plasmid from Burkolderia (Pseudomonas) cepacia G4 Appl. Environ. Microb. 61 1352–1356 Occurrence Handle1:CAS:528:DyaK2MXksl2ls7w%3D
K Solomon D Baker R Richards R Dixon S Klaine R Point R Kendall C Weisskopf J Giddings J Giesy L J Hall W Williams (1996) ArticleTitleEcological risk assessment of atrazine in North American surface waters Environ. Toxicol. Chem. 15 31–76 Occurrence Handle1:CAS:528:DyaK28XhvF2lsQ%3D%3D Occurrence Handle10.1897/1551-5028(1996)015<0031:ERAOAI>2.3.CO;2
D Smith S Alvey D E Crowley (2005) ArticleTitleCooperative catabolic pathways within an atrazine-degrading enrichment culture from soil FEMS Microbiol Ecol. 53 265–273 Occurrence Handle16329946 Occurrence Handle1:CAS:528:DC%2BD2MXltFaku7s%3D Occurrence Handle10.1016/j.femsec.2004.12.011
N Spliid B Koppen (1998) ArticleTitleOccurrence of pesticides in Danish shallow ground water Chemosphere 37 1307–1316 Occurrence Handle9734322 Occurrence Handle1:CAS:528:DyaK1cXlsVams7g%3D Occurrence Handle10.1016/S0045-6535(98)00128-3
L C Strong C Rosendahl G Johnson M J Sadowsky L P Wackett (2002) ArticleTitle Arthrobacter aurescens TC1 metabolizes diverse s-triazine ring compounds Appl. Environ. Microb. 68 5973–5980 Occurrence Handle1:CAS:528:DC%2BD38Xptlartrw%3D Occurrence Handle10.1128/AEM.68.12.5973-5980.2002
J Thompson T Gibson F Plewniak F Jeanmougin D Higgins (1997) ArticleTitleThe ClustalX Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools Nucl. Acids Res. 24 4876–4882 Occurrence Handle10.1093/nar/25.24.4876
I Tomasi I Artaud Y Bertheau D Mansuy (1995) ArticleTitleMetabolism of polychlorinated phenols by Pseudomonas cepacia AC1100; determination of the first two steps and specific inhibitory effect of methimazole J. Bacteriol. 177 307–311 Occurrence Handle7529225 Occurrence Handle1:CAS:528:DyaK2MXjtFOqs70%3D
E Topp W W Mulbry H Zhu S M Nour D Cuppels (2000a) ArticleTitleCharacterization of s-triazine herbicide metabolism by a Nocardioides sp. isolated from agricultural soils Appl. Environ. Microb. 66 3134–3141 Occurrence Handle1:CAS:528:DC%2BD3cXlsFaktbs%3D Occurrence Handle10.1128/AEM.66.8.3134-3141.2000
E Topp H Zhu S M Nour S Houot M Lewis D Cuppels (2000b) ArticleTitleCharacterization of an atrazine-degrading Pseudaminobacter sp. isolated from Canadian and French agricultural soils Appl. Environ. Microb. 66 2773–2782 Occurrence Handle1:CAS:528:DC%2BD3cXkslKlsL4%3D Occurrence Handle10.1128/AEM.66.7.2773-2782.2000
T Vallaeys L Courde C McGowan A D Wright R R Fulthorpe (1999) ArticleTitlePhylogenetic analyses indicate independent recruitment of diverse gene cassettes during assemblage of the 2,4-D catabolic pathway FEMS Microbiol. Ecol. 28 373–382 Occurrence Handle1:CAS:528:DyaK1MXit1Wru70%3D
A Yassir C Rieu G Soulas (1998) ArticleTitleMicrobial N-dealkylation of atrazine: effect of exogenous organic substrates and behaviour of the soil microflora Pest. Sci. 54 75–82 Occurrence Handle1:CAS:528:DyaK1cXmtFCqsLw%3D Occurrence Handle10.1002/(SICI)1096-9063(199809)54:1<75::AID-PS784>3.0.CO;2-3
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Martin-Laurent, F., Barrès, B., Wagschal, I. et al. Impact of the Maize Rhizosphere on the Genetic Structure, the Diversity and the Atrazine-degrading Gene Composition of Cultivable Atrazine-degrading Communities. Plant Soil 282, 99–115 (2006). https://doi.org/10.1007/s11104-005-5316-3
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/s11104-005-5316-3