Abstract
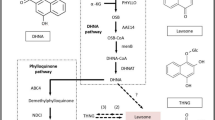
Fritillariae Bulbus are the most commonly used antitussive and edible herbs in China. Based on UPLC-QTOF-MS and UPLC-QQQ-MS, the validated MRM-based non-targeted quantitative method was applied to determinate the contents of 48 Fritillaria alkaloids (FAs) in three Fritillaria species (F. thunbergii Miq., F. unibracteata and F. ussuriensis). The RNA-Seq results showed that gene transcript levels have different expression patterns in three Fritillaria species. Based on transcriptome data, the full-length cDNA sequences of squalene epoxidase gene were cloned and characterized. Natural evolution of squalene epoxidase genes resulted in four mutations (C236R, M489L, G510A and K517R) in three Fritillaria species. Molecular docking analysis showed that the 236 residue is located inside the pocket and the binding center while other three residues are located on the surface of the protein. Functional verification indicated the mutations of SQE (C236R) could effectively increase the activity of SQE and obtain higher yield of 2,3-oxidosqualene in recombinant yeast. And the mutations of SQE (M489L and G510A), which increased the hydrophobicity of the protein surface, could also enhance the activity of SQE. This study provides major insights into the metabolites differentiation of FAs biosynthesis, and a firm foundation for the quality control and metabolic engineering of Fritillariae bulbus.
Key message
Deeper insights into the differentiation of metabolomics and transcriptomics and natural mutations of squalene epoxidase genes in three Fritillaria Bulbus.








Similar content being viewed by others
Abbreviations
- FAs:
-
Fritillaria Alkaloids
- Ft:
-
F. thunbergii Miq.
- Fun:
-
F. unibracteata
- Fus:
-
F. ussuriensis
- UPLC-QTOF-MS:
-
Ultra performance liquid chromatography-quadrupole time of flight-mass spectrometry
- UPLC-QQQ-MS:
-
Ultra performance liquid chromatography-triple quadrupole-mass spectrometry
- IPP:
-
Isopentenyl diphosphate
- DMAPP:
-
Dimethylallyl diphosphate
- IDI:
-
Isopentenyl diphosphate isomerase
- FPS:
-
Farnesyl diphosphate synthase
- SQS:
-
Squalene synthase
- SQE:
-
Squalene epoxidase
- CAS:
-
Cycloartenol synthase
- HMGR:
-
3-Hydroxy-3-methylglutaryl-CoA reductase
- MK:
-
Mevalonate kinase
- MDD:
-
Mevalonate pyrophosphate decarboxylase
References
Alseekh S, Tohge T, Wendenberg R, Scossa F, Omranian N, Li J et al (2015) Identification and mode of inheritance of quantitative trait loci for secondary metabolite abundance in tomato. Plant Cell 27:485–512
Benveniste P (2004) Biosynthesis and accumulation of sterols. Annu Rev Plant Biol 55:429–457
Brachmann CB, Davies A, Cost GJ, Caputo E, Li JC, Hieter P, Boeke JD (1998) Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115–132
Burke D, Dawson D, Stearns TIM (2000) Methods in yeast genetics: a Cold Spring Harbor Laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Cardenas PD, Sonawane PD, Pollier J, Vanden Bossche R, Dewangan V, Weithorn E et al (2016) GAME9 regulates the biosynthesis of steroidal alkaloids and upstream isoprenoids in the plant mevalonate pathway. Nat Commun 7:10654
Dai ZB, Liu Y, Huang LQ, Zhang XL (2012) Production of miltiradiene by metabolically engineered Saccharomyces cerevisiae. Biotechnol Bioeng 109:2845–2853
Dai ZB, Liu Y, Zhang X, Shi MY, Wang BB, Wang D et al (2013) Metabolic engineering of Saccharomyces cerevisiae for production of ginsenosides. Metab Eng 20:146–156
Donald KA, Hampton RY, Fritz IB (1997) Effects of overproduction of the catalytic domain of 3-hydroxy-3-methylglutaryl coenzyme A reductase on squalene synthesis in Saccharomyces cerevisiae. Appl Environ Microb 63:3341–3344
Forli S, Botta M (2007) Lennard-Jones potential and dummy atom settings to overcome the AUTODOCK limitation in treating flexible ring systems. J Chem Inf Model 47:1481–1492
Gietz RD, Schiestl RH, Willems AR, Woods RA (2010) Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11:355–360
Itkin M, Heinig U, Tzfadia O, Bhide AJ, Shinde B, Cardenas PD et al (2013) Biosynthesis of antinutritional alkaloids in solanaceous crops is mediated by clustered genes. Science 341:175–179
Jiang Y, Li P, Li HJ, Yu H (2006) New steroidal alkaloids from the bulbs of Fritillaria puqiensis. Steroids 71:843–848
Li HJ, Jiang Y, Li P (2007) Chemistry, bioactivity and geographical diversity of steroidal alkaloids from the Liliaceae family. Nat Prod Rep 38:735–752
Lin G, Li P, Li SL, Chan SW (2001) Chromatographic analysis of Fritillaria, isosteroidal alkaloids, the active ingredients of Beimu, the antitussive traditional Chinese medicinal herb. J Chromatogr A 935:321–338
Lin BQ, Ji H, Li P, Jiang Y, Fang W (2006) Selective antagonism activity of alkaloids from bulbs Fritillariae at muscarinic receptors: functional studies. Eur J Pharmacol 551:125–130
Pollier J, Vancaester E, Kuzhiumparambil U, Vickers CE, Vandepoele K, Goossens A et al (2019) A widespread alternative squalene epoxidase participates in eukaryote steroid biosynthesis. Nat Microbiol 4:226–233
Schaller H (2004) New aspects of sterol biosynthesis in growth and development of higher plants. Plant Physiol Biochem 42:465–476
Schäfer UA, Reed DW, Hunter DG, Yao KN, Weninger AM, Tsang EWT et al (1999) An example of intron junctional sliding in the gene families encoding squalene monooxygenase homologues in Arabidopsis thaliana and Brassica Napus. Plant Mol Biol 39:721–728
Schwede T, Kopp J, Guex N, Peitsch MC (2003) SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res 31:3381–3385
Song Y, Song Q, Liu Y, Li J, Wan JB, Wang YT et al (2017) Integrated work-flow for quantitative metabolome profiling of plants, Peucedani Radix as a case. Anal Chim Acta 953:40–47
Sun C, Sun YQ, Song JY, Li CJ, Li XW, Zhang XW et al (2011) Discovery of genes related to steroidal alkaloid biosynthesis in Fritillaria cirrhosa by generating and mining a dataset of expressed sequence tags (ESTs). J Med Plants Res 5:5307–5314
Wang L, Yao ZP, Li P, Chen SB, So PK, Shi ZQ et al (2016) Global detection and semi-quantification of Fritillaria alkaloids in Fritillariae Ussuriensis Bulbus by a non-targeted multiple reaction monitoring approach. J Sep Sci 39:287–295
Xin GZ, Hu B, Shi ZQ, Lam YC, Dong TTX, Li P et al (2014) Rapid identification of plant materials by wooden-tip electrospray ionization mass spectrometry and a strategy to differentiate the bulbs of Fritillaria. Anal Chim Acta 820:84–91
Zhao Q, Li R, Zhang Y, Huang KJ, Wang WG, Li J (2018) Transcriptome analysis reveals in vitro-cultured regeneration bulbs as a promising source for targeted Fritillaria cirrhosa steroidal alkaloid biosynthesis. 3 Biotech 8:191
Zhao FL, Bai P, Liu T, Li DS, Zhang XM, Lu WY et al (2016) Optimization of a Cytochrome P450 oxidation system for enhancing protopanaxadiol production in Saccharomyces cerevisiae. Biotechnol Bioeng 113(8):1787–1795
Zhou JL, Xin GZ, Shi ZQ, Ren MT, Qi LW, Li HJ et al (2010) Characterization and identification of steroidal alkaloids in Fritillaria species using liquid chromatography coupled with electrospray ionization quadrupole time-of-flight tandem mass spectrometry. J Chromatogr A 1217:7109–7712
Acknowledgements
We acknowledge Prof. Zhang Xueli for providing the plasmid of pRS313, and the Biotechnology Research Institute at the Chinese Academy of Agricultural Sciences for its assistance in original data processing and related bioinformatics analysis. This work was supported by the National Key R&D Program of China (2019YFC1711000), National Natural Science Foundation of China (Grant Nos. 81773872, 81322051 and 81973414), Natural Science Foundation of Jiangsu Province (Grant No. BK20191319), Fundamental Research Funds for the Central Universities (2632019ZD15), "Double First-Class" University project (CPU2018GY09) and “111” project (Grant No. B16046).
Author information
Authors and Affiliations
Contributions
PL, GZX and YJ conceived and designed this research. XL, LNZ and JFD investigated, analyzed data and wrote the manuscript. XYZ participated in the discussion of the results. HJL contributed to the evaluation and discussion of the results and manuscript revision.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Lu, X., Zhang, LN., Du, JF. et al. Comparative analysis and natural evolution of squalene epoxidase in three Fritillaria species. Plant Mol Biol 103, 705–718 (2020). https://doi.org/10.1007/s11103-020-01021-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-020-01021-y