Abstract
Key message
miR171a controls HAM1 functions within the protodermal cells of the embryo, and these controls are essential for normal embryogenesis in Arabidopsis.
Abstract
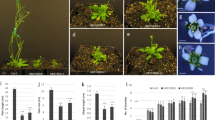
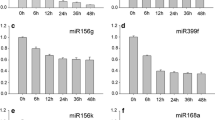
Arabidopsis thaliana miR171a is known to bind to and cleave mRNAs of three HAIRY MERISTEM (HAM) genes that encode members of the GRAS family transcriptional regulators. The molecular functions of the HAM genes are still being elucidated in Arabidopsis. However, detailed expression patterns of miR171a and the effects of the failure of miR171a to suppress HAM genes were unknown till now. Here, we show the detailed expression patterns of miR171a and HAM1 using green fluorescent protein and confocal scanning microscopy. Our observations revealed that miR171a was expressed in the surface cell layer of the embryo and shoot apical meristem, and it controlled HAM1 functions. To determine the impact of the failure of miR171a to suppress of HAM1, we introduced seven synonymous mutations into the miR171a target site of the HAM1 gene (modified HAM1, mHAM1) and generated transgenic plants that had mHAM1 driven by HAM1 native promoter. The mHAM1 transgenic plants showed organogenic defects. These results indicate that the control of HAM1 functions at the single-cell-layer level by miR171a is essential for proper organ formation in Arabidopsis.





Similar content being viewed by others
References
Achkar NP, Cambiagno DA, Manavella PA (2016) miRNA biogenesis: a dynamic pathway. Trends Plant Sci 21:1034–1044. https://doi.org/10.1016/j.tplants.2016.09.003
Aida M, Ishida T, Tasaka M (1999) Shoot apical meristem and cotyledon formation during Arabidopsis embryogenesis: interaction among the CUP-SHAPED COTYLEDON and SHOOT MERISTEMLESS genes. Development 126:1563–1570
Baker CC, Sieber P, Wellmer F, Meyerowitz EM (2005) The early extra petals1 mutant uncovers a role for MicroRNA miR164c in regulating petal number in Arabidopsis. Curr Biol 15:303–315. https://doi.org/10.1016/j.cub.2005.02.017
Bologna NG, Voinnet O (2014) The diversity, biogenesis, and activities of endogenous silencing small RNAs in Arabidopsis. Annu Rev Plant Biol 65:473–503. https://doi.org/10.1146/annurev-arplant-050213-035728
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743. https://doi.org/10.1046/j.1365-313x.1998.00343.x
Emery JF, Floyd SK, Alvarez J, Eshed Y, Hawker NP, Izhaki A, Baum SF, Bowman JL (2003) Radial patterning of Arabidopsis shoots by class IIIHD-ZIP and KANADI genes. Curr Biol 13:1768–1774. https://doi.org/10.1016/j.cub.2003.09.035
Engstrom EM, Andersen CM, Gumulak-Smith J, Hu J, Orlova E, Sozzani R, Bowman JL (2011) Arabidopsis homologs of the petunia hairy meristem gene are required for maintenance of shoot and root indeterminacy. Plant Physiol 155:735–750. https://doi.org/10.1104/pp.110.168757
Hardtke CS, Berleth T (1998) The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J 17:1405–1411. https://doi.org/10.1093/emboj/17.5.1405
Hibara K, Karim MR, Takada S, Taoka KI, Furutani M, Aida M, Tasaka M (2006) Arabidopsis CUP-SHAPED COTYLEDON3 regulates postembryonic shoot meristem and organ boundary formation. Plant Cell 18:2946–2957. https://doi.org/10.1105/tpc.106.045716
Huang J, Fujimoto M, Fujiwara M, Fukao Y, Arimura S, Tsutsumi N (2015) Arabidopsis dynamin-related proteins, DRP2A and DRP2B, function coordinately in post-Golgi trafficking. Biochem Biophys Res Commun 456:238–244. https://doi.org/10.1016/j.bbrc.2014.11.065
Karimi M, Inze D, Depicker A (2002) GATEWAY((TM)) vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7:193–195. https://doi.org/10.1016/s1360-1385(02)02251-3
Klepikova AV, Kasianov AS, Gerasimov ES, Logacheva MD, Penin AA (2016) A high resolution map of the Arabidopsis thaliana developmental transcriptome based on RNA-seq profiling. Plant J 88:1058–1070. https://doi.org/10.1111/tpj.13312
Kozomara A, Griffiths-Jones S (2014) miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 42:D68–D73. https://doi.org/10.1093/nar/gkt1181
Lee RC, Feinbaum RL, Ambros V (1993) The C. elegans heterochronic gene lin-4 encodes small rnas with antisense complementarity to lin-14. Cell 75:843–854. https://doi.org/10.1016/0092-8674(93)90529-y
Li C, Zhang BH (2016) MicroRNAs in control of plant development. J Cell Physiol 231:303–313. https://doi.org/10.1002/jcp.25125
Llave C, Xie Z, Kasschau KD, Carrington JC (2002) Cleavage of scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297:2053–2056. https://doi.org/10.1126/science.1076311
Mallory AC, Vaucheret H (2006) Functions of microRNAs and related small RNAs in plants. Nat Genet 38:S31–S36. https://doi.org/10.1038/ng1791
Manavella PA, Koenig D, Rubio-Somoza I, Burbano HA, Becker C, Weigel D (2013) Tissue-specific silencing of Arabidopsis SU(VAR)3–9 HOMOLOG8 by miR171a. Plant Physiol 161:805–812. https://doi.org/10.1104/pp.112.207068
McConnell JR, Barton MK (1998) Leaf polarity and meristem formation in Arabidopsis. Development 125:2935–2942
Meunier J, Lemoine F, Soumillon M, Liechti A, Weier M, Guschanski K, Hu HY, Khaitovich P, Kaessmann H (2013) Birth and expression evolution of mammalian microRNA genes. Genome Res 23:34–45. https://doi.org/10.1101/gr.140269.112
Miyashima S, Honda M, Hashimoto K, Tatematsu K, Hashimoto T, Sato-Nara K, Okada K, Nakajima K (2013) A comprehensive expression analysis of the Arabidopsis MICRORNA165/6 gene family during embryogenesis reveals a conserved role in meristem specification and a non-cell-autonomous function. Plant Cell Physiol 54:375–384. https://doi.org/10.1093/pcp/pcs188
Montes RAC, Rosas-Cardenas FD, De Paoli E, Accerbi M, Rymarquis LA, Mahalingam G et al. (2014) Sample sequencing of vascular plants demonstrates widespread conservation and divergence of microRNAs. Nat Commun. https://doi.org/10.1038/ncomms4722
Odat O, Gardiner J, Sawchuk MG, Verna C, Donner TJ, Scarpella E (2014) Characterization of an allelic series in the MONOPTEROS gene of Arabidopsis. Genesis 52:127–133. https://doi.org/10.1002/dvg.22729
Ohnishi T, Takanashi H, Mogi M, Takahashi H, Kikuchi S, Yano K et al (2011) Distinct gene expression profiles in egg and synergid cells of rice as revealed by cell type-specific microarrays. Plant Physiol 155:881–891. https://doi.org/10.1104/pp.110.167502
Raman S, Greb T, Peaucelle A, Blein T, Laufs P, Theres K (2008) Interplay of miR164, CUP-SHAPED COTYLEDON genes and LATERAL SUPPRESSOR controls axillary meristem formation in Arabidopsis thaliana. Plant J 55:65–76. https://doi.org/10.1111/j.1365-313X.2008.03483.x
Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP (2002) MicroRNAs in plants. Genes Dev 16:1616–1626. https://doi.org/10.1101/gad.1004402
Rodriguez RE, Schommer C, Palatnik JF (2016) Control of cell proliferation by microRNAs in plants. Curr Opin Plant Biol 34:68–76. https://doi.org/10.1016/j.pbi.2016.10.003
Scheres B, Wolkenfelt H, Willemsen V, Terlouw M, Lawson E, Dean C, Weisbeek P (1994) Embryonic origin of the Arabidopsis primary root and root-meristem initials. Development 120:2475–2487
Schulze S, Schafer BN, Parizotto EA, Voinnet O, Theres K (2010) LOST MERISTEMS genes regulate cell differentiation of central zone descendants in Arabidopsis shoot meristems. Plant J 64:668–678. https://doi.org/10.1111/j.1365-313X.2010.04359.x
Stuurman J, Jaggi F, Kuhlemeier C (2002) Shoot meristem maintenance is controlled by a GRAS-gene mediated signal from differentiating cells. Genes Dev 16:2213–2218. https://doi.org/10.1101/gad.230702
Szemenyei H, Hannon M, Long JA (2008) TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319:1384–1386. https://doi.org/10.1126/science.1151461
Wang L, Mai YX, Zhang YC, Luo Q, Yang HQ (2010) MicroRNA171c-targeted SCL6-II, SCL6-III, and SCL6-IV genes regulate shoot branching in Arabidopsis. Mol Plant 3:794–806. https://doi.org/10.1093/mp/ssq042
Willmann MR, Poethig RS (2007) Conservation and evolution of miRNA regulatory programs in plant development. Curr Opin Plant Biol 10:503–511. https://doi.org/10.1016/j.pbi.2007.07.004
Zhou GK, Kubo M, Zhong RQ, Demura T, Ye ZH (2007) Overexpression of miR165 affects apical meristem formation, organ polarity establishment and vascular development in Arabidopsis. Plant Cell Physiol 48:391–404. https://doi.org/10.1093/pcp/pcm008
Zhou Y, Liu X, Engstrom EM, Nimchuk ZL, Pruneda-Paz JL, Tarr PT, Yan A, Kay SA, Meyerowitz EM (2015) Control of plant stem cell function by conserved interacting transcriptional regulators. Nature 517:377–380. https://doi.org/10.1038/nature13853
Funding
This research was supported by Grants-in-Aid for Scientific Research on Priority Areas (Grant 18075005 to N.T.) and Exploratory Research (Grant 26660004 to H.T.) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Author information
Authors and Affiliations
Contributions
HT designed the study, and wrote the initial draft of the manuscript. MM contributed to constructing expression vectors. HS and YH contributed to analysis and observation of transgenic plants. TO and NT helped supervise the project. All authors discussed the results and contributed to the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing or financial interests.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Takanashi, H., Sumiyoshi, H., Mogi, M. et al. miRNAs control HAM1 functions at the single-cell-layer level and are essential for normal embryogenesis in Arabidopsis. Plant Mol Biol 96, 627–640 (2018). https://doi.org/10.1007/s11103-018-0719-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-018-0719-8