Abstract
Key message
Related motifs strongly increase gene expression when added to an intron located in coding sequences.
Abstract
Many introns greatly increase gene expression through a mechanism that remains elusive. An obstacle to understanding intron-mediated enhancement (IME) has been the difficulty of locating the specific intron sequences responsible for boosting expression because they are redundant, dispersed, and degenerate. Previously we used the IMEter algorithm in two independent ways to identify two motifs (CGATT and TTNGATYTG) that are candidates for involvement in IME in Arabidopsis. Here we show that both motifs are sufficient to increase expression. An intron that has little influence on expression was converted into one that increased mRNA accumulation 24-fold and reporter enzyme activity 40-fold relative to the intronless control by introducing 11 copies of the more active TTNGATYTG motif. This degree of stimulation is twice as large as that of the strongest of 15 natural introns previously tested in the same reporter gene. Even though the CGATT and TTNGATYTG motifs each increased expression, and CGATT matches the NGATY core of the longer motif, combining the motifs to make TTCGATTTG reduced the stimulating ability of the TTNGATYTG motif. Additional substitutions were used to test the contribution to IME of other residues in the TTNGATYTG motif. The verification that these motifs are active in IME will improve our ability to predict the stimulating ability of introns, to engineer any intron to increase expression to a desired level, and to explore the mechanism of IME by seeking factors that might interact with these sequences.




Similar content being viewed by others
References
Akua T, Shaul O (2013) The Arabidopsis thaliana MHX gene includes an intronic element that boosts translation when localized in a 5′ UTR intron. J Exp Bot 64:4255–4270
Bailey TL, Williams N, Misleh C, Li WW (2006) MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res 34:W369–W373
Bianchi M, Crinelli R, Giacomini E, Carloni E, Magnani M (2009) A potent enhancer element in the 5′-UTR intron is crucial for transcriptional regulation of the human ubiquitin C gene. Gene 448:88–101
Buchman AR, Berg P (1988) Comparison of intron-dependent and intron-independent gene expression. Mol Cell Biol 8:4395–4405
Callis J, Fromm M, Walbot V (1987) Introns increase gene expression in cultured maize cells. Genes Dev 1:1183–1200
Chodavarapu RK et al (2010) Relationship between nucleosome positioning and DNA methylation. Nature 466:388–392
Chung BY, Simons C, Firth AE, Brown CM, Hellens RP (2006) Effect of 5′UTR introns on gene expression in Arabidopsis thaliana. BMC Genomics 7:120
Clancy M, Hannah LC (2002) Splicing of the Maize Sh1 first intron is essential for enhancement of gene expression, and a T-rich motif increases expression without affecting splicing. Plant Physiol 130:918–929
Clancy M, Vasil V, Hannah LC, Vasil IK (1994) Maize Shrunken-1 intron and exon regions increase gene expression in maize protoplasts. Plant Sci 98:151–161
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Deyholos MK, Sieburth LE (2000) Separable whorl-specific expression and negative regulation by enhancer elements within the AGAMOUS second intron. Plant Cell 12:1799–1810
Dieci G, Fiorino G, Castelnuovo M, Teichmann M, Pagano A (2007) The expanding RNA polymerase III transcriptome. Trends Genet 23:614–622
Donath M, Mendel R, Cerff R, Martin W (1995) Intron-dependent transient expression of the maize GapA1 gene. Plant Mol Biol 28:667–676
Down TA, Hubbard TJP (2005) NestedMICA: sensitive inference of over-represented motifs in nucleic acid sequence. Nucleic Acids Res 33:1445–1453
Duncker BP, Davies PL, Walker VK (1997) Introns boost transgene expression in Drosophila melanogaster. Mol Gen Genet 254:291–296
Eamens A, Wang MB, Smith NA, Waterhouse PM (2008) RNA silencing in plants: yesterday, today, and tomorrow. Plant Physiol 147:456–468
Emami S, Arumainayagam D, Korf I, Rose AB (2013) The effects of a stimulating intron on the expression of heterologous genes in Arabidopsis thaliana. Plant Biotechnol J 11:555–563
Gallegos JE, Rose AB (2015) The enduring mystery of intron-mediated enhancement. Plant Sci 237:8–15
Gruss P, Lai CJ, Dhar R, Khoury G (1979) Splicing as a Requirement for Biogenesis of Functional 16 S Messenger-RNA of Simian Virus-40. Proc Natl Acad Sci USA 76:4317–4321
Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27:297–300
Jefferson RA (1987) Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Rep 5:387–405
Jeon JS, Lee S, Jung KH, Jun SH, Kim C, An G (2000) Tissue-preferential expression of a rice alpha-tubulin gene, OsTubA1, mediated by the first intron. Plant Physiol 123:1005–1014
Jeong YM, Mun JH, Lee I, Woo JC, Hong CB, Kim SG (2006) Distinct roles of the first introns on the expression of Arabidopsis profilin gene family members. Plant Physiol 140:196–209
Juneau K, Miranda M, Hillenmeyer ME, Nislow C, Davis RW (2006) Introns regulate RNA and protein abundance in yeast. Genetics 174:511–518
Le Hir H, Nott A, Moore MJ (2003) How introns influence and enhance eukaryotic gene expression. Trends Biochem Sci 28:215–220
Lescot M et al (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30:325–327
Lu S, Cullen BR (2003) Analysis of the stimulatory effect of splicing on mRNA production and utilization in mammalian cells. RNA 9:618–630
Luehrsen KR, Walbot V (1994) Addition of A- and U-rich sequence increases the splicing efficiency of a deleted form of a maize intron. Plant Mol Biol 24:449–463
Matsumoto K, Wassarman KM, Wolffe AP (1998) Nuclear history of a pre-mRNA determines the translational activity of cytoplasmic mRNA. EMBO J 17:2107–2121
Moabbi AM, Agarwal N, El Kaderi B, Ansari A (2012) Role for gene looping in intron-mediated enhancement of transcription. Proc Natl Acad Sci U S A 109:8505–8510
Nagaya S, Kato K, Ninomiya Y, Horie R, Sekine M, Yoshida K, Shinmyo A (2005) Expression of randomly integrated single complete copy transgenes does not vary in Arabidopsis thaliana. Plant Cell Physiol 46:438–444
Niu DK, Yang YF (2011) Why eukaryotic cells use introns to enhance gene expression: splicing reduces transcription-associated mutagenesis by inhibiting topoisomerase I cutting activity. Biol Direct 6:24
Nott A, Le Hir H, Moore MJ (2004) Splicing enhances translation in mammalian cells: an additional function of the exon junction complex. Genes Dev 18:210–222
Okkema PG, Harrison SW, Plunger V, Aryana A, Fire A (1993) Sequence requirements for myosin gene expression and regulation in Caenorhabditis elegans. Genetics 135:385–404
Palmiter RD, Sandgren EP, Avarbock MR, Allen DD, Brinster RL (1991) Heterologous introns can enhance expression of transgenes in mice. Proc Natl Acad Sci USA 88:478–482
Parra G, Bradnam K, Rose AB, Korf I (2011) Comparative and functional analysis of intron-mediated enhancement signals reveals conserved features among plants. Nucleic Acids Res 39:5328–5337
Rose AB (2002) Requirements for intron-mediated enhancement of gene expression in Arabidopsis. RNA 8:1444–1453
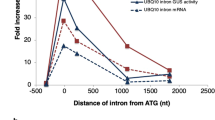
Rose AB (2004) The effect of intron location on intron-mediated enhancement of gene expression in Arabidopsis. Plant J 40:744–751
Rose AB, Elfersi T, Parra G, Korf I (2008) Promoter-proximal introns in Arabidopsis thaliana are enriched in dispersed signals that elevate gene expression. Plant Cell 20:543–551
Rose AB, Emami S, Bradnam K, Korf I (2011) Evidence for a DNA-based mechanism of intron-mediated enhancement front. Plant Sci 2:98
Schubert D, Lechtenberg B, Forsbach A, Gils M, Bahadur S, Schmidt R (2004) Silencing in Arabidopsis T-DNA transformants: the predominant role of a gene-specific RNA sensing mechanism versus position effects. Plant Cell 16:2561–2572
Schwartz S, Meshorer E, Ast G (2009) Chromatin organization marks exon–intron structure. Nat Struct Mol Biol 16:990–995
Segal E et al (2006) A genomic code for nucleosome positioning. Nature 442:772–778
Snowden KC, Buchholz WG, Hall TC (1996) Intron position affects expression from the tpi promoter in rice. Plant Mol Biol 31:689–692
Tilgner H, Nikolaou C, Althammer S, Sammeth M, Beato M, Valcarcel J, Guigo R (2009) Nucleosome positioning as a determinant of exon recognition. Nat Struct Mol Biol 16:996–1001
Wiegand HL, Lu S, Cullen BR (2003) Exon junction complexes mediate the enhancing effect of splicing on mRNA expression. Proc Natl Acad Sci USA 100:11327–11332
Yilmaz A, Mejia-Guerra MK, Kurz K, Liang X, Welch L, Grotewold E (2011) AGRIS: the Arabidopsis gene regulatory information server, an update. Nucleic Acids Res 39:D1118–D1122
Acknowledgments
We thank Dr. Neil Willits for statistical analysis, and Dr. Lesilee Rose and Jenna Gallegos for helpful comments on the manuscript. This work was supported in part by the United States Department of Agriculture, Grant Number 2006-35301-17072.
Author contributions
A.R designed and carried out most of the experiments, and wrote the manuscript. A.C. and N.K. assisted in the experiments. I.K. provided bioinformatic guidance in choosing motif mutations to test.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
11103_2016_516_MOESM2_ESM.pptx
Supplementary Fig. 1 The ability of introns containing different portions of the UBQ10 intron to increase expression. The UBQ10 intron and derivatives with various sections deleted (triangles), or hybrid introns composed of different parts of the UBQ10 and COR15a introns (circles) were inserted at the same location of the TRP1:GUS reporter gene. The effect of each intron on mRNA accumulation relative to an intronless control in single-copy transgenic lines is plotted as a function of the total number of nucleotides of UBQ10 sequence present in the intron. Compiled from data for introns described in (Rose et al. 2008) and (Parra et al. 2011) (PPTX 47 KB)
Rights and permissions
About this article
Cite this article
Rose, A.B., Carter, A., Korf, I. et al. Intron sequences that stimulate gene expression in Arabidopsis . Plant Mol Biol 92, 337–346 (2016). https://doi.org/10.1007/s11103-016-0516-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-016-0516-1