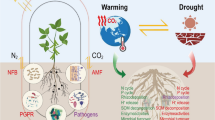
Abstract
The rhizosphere microbiome is pivotal for plant health and growth, providing defence against pests and diseases, facilitating nutrient acquisition and helping plants to withstand abiotic stresses. Plants can actively recruit members of the soil microbial community for positive feedbacks, but the underlying mechanisms and plant traits that drive microbiome assembly and functions are largely unknown. Domestication of plant species has substantially contributed to human civilization, but also caused a strong decrease in the genetic diversity of modern crop cultivars that may have affected the ability of plants to establish beneficial associations with rhizosphere microbes. Here, we review how plants shape the rhizosphere microbiome and how domestication may have impacted rhizosphere microbiome assembly and functions via habitat expansion and via changes in crop management practices, root exudation, root architecture, and plant litter quality. We also propose a “back to the roots” framework that comprises the exploration of the microbiome of indigenous plants and their native habitats for the identification of plant and microbial traits with the ultimate goal to reinstate beneficial associations that may have been undermined during plant domestication.
Similar content being viewed by others
Introduction
Plants rely on their rhizosphere microbiome for functions and traits related to plant growth, development and health (Berendsen et al. 2012; Mendes et al. 2013). Members of the rhizosphere microbiome harbour a range of beneficial properties contributing to nutrient acquisition, enhanced stress tolerance, protection against soil borne pathogens and host immune regulation (Berendsen et al. 2012; Bakker et al. 2013; Mendes et al. 2013; Turner et al. 2013a; Berg et al. 2014; Lakshmanan et al. 2014). In this context, Cook et al. (1995) postulated that natural selection resulted in only few examples of plant genetic resistance against belowground pathogens and that plant rely, in part, on the natural defence provided by rhizosphere microorganisms. This is the case for natural disease suppressive soils where specific microbial consortia protect the host from infection (Mendes et al. 2011). Assuming that plants depend, at least in part, on the rhizosphere microbiome as a product of natural selection, modern cultivars of crop plants may have lost some of the traits needed to recruit host-specific root microbiota as compared to their wild relatives, which are genetically more diverse and adapted to pre-agricultural soils (Wissuwa et al. 2009; Bulgarelli et al. 2013). Whether the ability of crop plants to recruit beneficial rhizosphere microbes is undermined by plant domestication and plant breeding is not well known to date. In this review, we discuss the potential influence of plant domestication on rhizosphere microbiome assembly and function, focusing on how domestication may have impacted the ability of modern crops to establish beneficial interactions with the rhizosphere microbiome. Finally, we propose a framework for identification and recovery of beneficial plant–microbe interactions to meet the need for a more sustainable and productive agriculture.
Plant domestication: changes and trade-offs
One of the biggest accomplishments in human history has been the domestication of plants, providing a more continuous food supply and promoting the conformation of sedentary agricultural groups (Purugganan and Fuller 2009). The process of plant domestication involves selection, modification and adoption of wild plants species with useful characteristics for human use (Gepts 2004). The first changes commonly associated with plant domestication were a large seed size, loss of seed dispersal mechanisms, and determinate growth and apical dominance (Gross and Olsen 2010). Other changes comprise the loss of seed dormancy, decrease of bitter substances in edible structures and changes in photoperiod sensitivity (Purugganan and Fuller 2009). Domestication also led to a reduction in genetic diversity of plant cultivars as shown for common bean (Bitocchi et al. 2012, 2013), rice (Ram et al. 2007) and wheat (Haudry et al. 2007). Genes associated with desirable phenotypes underwent a diversity loss because only the desired alleles were spread in the subsequent progenies, whilst unwanted diversity of the same allele was inadvertently suppressed (Doebley et al. 2006). In addition, genomic regions next to the target genes suffered selective sweeps as was shown for the adjacent regions of the Y1 phytoene synthase gene for endosperm colour in maize (Palaisa et al. 2004) and of the Waxy granule-bound starch synthase gene for amylose synthesis in rice (Olsen et al. 2006). Thus, a possible side effect of plant domestication is the loss of traits neglected during human selection. In a recent review, Chen et al. (2015) indicated that the ability of plants to deal with herbivorous insects is undermined in domesticated crops, in part as a consequence of changes in morphological traits and in levels of secondary metabolites, which make domesticated plants a better resource for insects as compared to wild relatives. Chen et al. (2015) further highlighted that domestication led to lower levels of volatile emissions as compared to wild relatives, which in turn may affect the attraction of natural enemies. Whether plant traits needed to recruit and sustain beneficial microbial populations in the rhizosphere was also negatively impacted remains to be elucidated.
Many of the changes in plant traits during domestication were accompanied by progressive changes in the environment and management practices (Fig. 1). Hence, plant domestication associated with anthropogenic interference to sustain high yields led to low self-support production systems with an increased need for external inputs such as chemical pesticides and fertilizers to overcome problems related to pests and diseases, vulnerability to abiotic stress and nutrient depletion (Matson et al. 1997). Moreover, the transition from natural to agricultural systems may have hampered beneficial interactions between plants and microbes due to loss of soil microbial diversity. For instance, it was shown that long-term nitrogen fertilization resulted in the evolution of less-mutualistic rhizobia, providing fewer benefits to the host (Weese et al. 2015). Nitrogen amendments have also been shown to suppress soil respiration and microbial biomass, promoting copiotrophs such as Actinobacteria and Firmicutes while reducing the abundance of oligotrophs such as Acidobacteria and Verrucomicrobia (Ramirez et al. 2012). This was substantiated by Rodrigues et al. (2013) who showed that conversion of the Amazon rainforest to agriculture resulted in biotic homogenization of soil bacterial communities and reduction of microbial diversity. Fierer et al. (2013) further showed that in a native tallgrass prairie ecosystem, bacterial communities did not resemble those harboured by the surrounding cultivated soils where Verrucomicrobia represented more than 50 % of the 16S rRNA sequences identified. Also soil attributes can be affected by plant domestication, which in turn influence the soil microbial community composition. García-Palacios et al. (2013) demonstrated, in microbial-rich and microbial-poor soils, that plant domestication increased litter quality, resulting in lower C:N ratio and higher NO3 availability. In addition to changes in the production systems, domesticated lineages experienced range expansions far beyond their centres of origin due to human migrations and trade (Purugganan and Fuller 2009). Hence, the lack of a co-evolutionary trajectory between plants, microbial communities and pathogens in dissimilar agricultural landscapes, made human interventions even more critical to maintain a healthy and productive crop (Fig. 2).
In this image, the natural ecosystem is illustrated by the native Amazon rainforest (background) that was converted to a modern crop system (front). This conversion leads to changes in the environment and use of management techniques ultimately impacting the rhizosphere microbiome assembly and functions (Photo by L. W. Mendes)
Effect of plant genotype on rhizosphere microbiome assembly
Plants can modulate their rhizosphere microbiome in a host-dependent way. Each plant species promotes a particular set of rhizosphere microbes (Haichar et al. 2008; Turner et al. 2013b; Ofek et al. 2014). With an increase in the phylogenetic distance between plant species also differences in the composition of their rhizosphere microbial assemblages appear to increase (Wieland et al. 2001; Pongsilp et al. 2012; Bouffaud et al. 2014). Not only different plant species, but also different genotypes of the same species may differ in their rhizosphere microbiome composition. For example, Weinert et al. (2011) showed for three different potato cultivars that a portion of the detected OTUs was cultivar-specific and that the Streptomycetaceae responded in a cultivar-dependent manner. Similar cultivar-dependent effects were observed for the rhizobacterial communities in the rhizosphere of young potato plants (Inceoglu et al. 2011). In a recent study with 27 modern maize inbred lines, grown in five field environments, Peiffer et al. (2013) showed that OTU richness was affected by maize genotypes and that the variation in β-diversity was partially explained by the maize genotype. Similarly, in a study with different barley genotypes, Bulgarelli et al. (2015) found that the host genotype accounts for approximately 5.7 % of the variance in the rhizosphere microbiome composition. In sweet potato (Ipomoea batatas), Sphingobium, Pseudomonas, Acinetobacter, Stenotrophomonas, and Chryseobacterium were enriched on the low starch genotype as compared to two high starch genotypes (Marques et al. 2014). Next to genotype-specific effects, also the plant developmental stage is a strong driver shaping the rhizobacterial community structure. In soybean, Bradyrhizobium, Bacillus and Stenotrophomonas were more abundant at the flowering stage as compared to vegetative and mature stages (Sugiyama et al. 2014a). For fungal communities, however, no significant effects of the soybean growth stage were detected (Sugiyama et al. 2014b). These effects, however, are not general as some studies highlighted a stronger selective rhizosphere effect at young plant growth stages (Gomes et al. 2001; Jin et al. 2009; Micallef et al. 2009a; Xu et al. 2009; Chaparro et al. 2014) whilst other studies documented stronger selective effects on the microbiome at flowering (Smalla et al. 2001; Inceoglu et al. 2010).
Plant root exudates and the recruitment of beneficial microbes
Plants actively release exudates, volatile compounds, border cells and polymers into the soil, a process referred to as rhizodeposition (Jones et al. 2009). Root exudates are composed of low-molecular weight compounds, i.e. sugars, amino acids, organic acids, phenolics, secondary metabolites, and high-molecular weight compounds like proteins and mucilage (Badri and Vivanco 2009). For more details on the chemical diversity of compounds in the rhizosphere we refer to other reviews (Bais et al. 2006; Badri et al. 2009; Moe 2013; Weston and Mathesius 2013; Baetz and Martinoia 2014; Haichar et al. 2014).
Root exudates may impact the functioning of the microbial community. For instance, in soil amended with maize mucilage a higher production of N2O was recorded than in non-amended soil (Mounier et al. 2004). Likewise, additions of artificial root exudates (ARE) to a soil microcosm, mimicking maize exudates, promoted nitrate reduction and denitrification (Henry et al. 2008). A study with eight Arabidopsis thaliana accessions revealed that each accession released a particular set of exudate compounds and that each accession had a distinct rhizobacterial community composition based on RISA and 16S-TRFLP analyses (Micallef et al. 2009b). Some root exudates may impact the microbial community structure to a greater extent than other compounds as was shown for organic acids with a 10–22 fold increase in the bacterial taxa while sugars showed only a 2.5 fold increase (Shi et al. 2011). When Arabidopsis exudates collected from 18 to 21 days old plants were applied to a fallow soil, phenolic compounds had a significant positive correlation with the highest number of bacterial OTUs (742) whereas lower number of OTUs were found for amino acids (319), sugar alcohols (166), and sugars (161) (Badri et al. 2013). Root exudates such as flavonoids or strigolactones play key roles in symbiotic relationships between plants and rhizobia, mycorrhiza and also parasites (Jones et al. 2004; Bouwmeester et al. 2007; Bednarek et al. 2010; Wang et al. 2012; Haichar et al. 2014). Likewise, root exudates may impact specific groups of beneficial bacteria referred as plant growth promoting rhizobacteria (PGPR). For example, Pseudomonas fluorescens WCS365, a rhizosphere competent colonizer of tomato roots, was chemotactically attracted towards malic acid and citric acid exuded by tomato roots (de Weert et al. 2002). Also beneficial rhizobacterium Bacillus subtilis FB17 exhibited a positive chemotactic response towards L-malic acid. Interestingly, infection of Arabidopsis leaves with Pseudomonas syringae pv tomato induced an increased secretion of malic acid from the roots, promoting the colonization and biofilm formation by strain FB17 (Rudrappa et al. 2008). Furthermore, foliar pathogens or foliar treatment with microbe-associated molecular patterns (MAMPs) of Arabidopsis leaves promoted the expression of the root malic acid (MA) transporter (ALMT1), stimulating the colonization by B. subtilis strain FB17 (Lakshmanan et al. 2012). Malic acid and citric acid exuded by watermelon roots were shown to induce motility and root colonization by the PGPR Paenibacillus polymyxa SQR-21 (Ling et al. 2011). Similar effects of malic acid and citric acid were found for chemotaxis and biofilm formation by Bacillus amyloliquefaciens SQR9 in cucumber and for fumaric acid in promoting colonization of banana roots by B. subtilis N11 (Zhang et al. 2014). Also other compounds found in root exudates may recruit beneficial bacteria. The aromatic compound 2,4-dihydroxy-7-methoxy-2H-1,4-benzoxazin-3(4H)-one (DIMBOA) exuded by maize roots, showed a chemoattractant effect on and an increased root colonization by Pseudomonas putida KT2440 (Neal et al. 2012). Finally, plant derived compounds may also have an effect on the expression of bacterial antifungal biosynthetic genes. For instance, the expression of phlA and pltA genes in P. fluorescens CHA0, involved in the biosynthesis of the antifungal compounds 2,4-diacetylphloroglucinol (DAPG) and pyoluteorin (PLT) respectively, was induced or repressed by 40 different plant-derived compounds, including several plant phenolics and pectin (de Werra et al. 2011). Similarly, phenolic and organic acids exuded by barley plants infected with the fungus Pythium ultimum induced the expression of the phlA gene of P. fluorescens CHA0, presumably as a plant systemic response to deal with the pathogen (Jousset et al. 2011). Also Zea mays subsp. parviglumis and European maize lines emitted the volatile sesquiterpene (E)-β-caryophyllene via the roots attracting an entomopathogenic nematode in response to insect attack; North American lines failed to release this compound probably as a consequence of the breeding process (Rasmann et al. 2005; Köllner et al. 2008).
These results exemplify the potential of plants to recruit and activate, via specific components in root exudates, beneficial members of the rhizosphere microbiome. However, plant exudates may also exert a negative effect on belowground communities. In a study evaluating the effect of the invasive weed Centaurea maculosa on the composition of arbuscular mycorrhizal fungal (AMF) communities, the abundance and diversity of AMF was reduced compared to native grassland samples (Mummey and Rillig 2006). The same deleterious effect of C. maculosa was also shown for the overall soil fungal community. Broz et al. (2007) further observed that high density weed populations had a reduced fungal biomass and diversity as compared to low density weed populations mixed with native species. Badri and Vivanco (2009) suggested that root exudates released by invasive weeds disrupt the indigenous microbial communities probably through an antimicrobial effect. Although the available results are still limited, fragmentary and not conclusive, one may assume that plant domestication can lead to changes in root exudation profiles and thereby impact on the rhizosphere microbiome composition and function (Fig. 2).
Effect of plant domestication on belowground interactions
Undermined mycorrhizal symbiosis
The effect of domestication and plant breeding on belowground interactions with soil microorganisms was addressed by pioneering studies with wheat evaluating the ability of ancestors, landraces and modern genotypes to sustain mycorrhizal symbiosis (Kapulnik and Kushnir 1991). The mycorrhizal dependence (MD), i.e. the degree of dependence on mycorrhizal symbiosis for maximum plant growth and yield, was determined for wild and cultivated wheat genotypes. The results showed that a diploid wheat ancestor, Triticum tauschii, displayed a higher MD compared to tetraploid or modern hexaploid wheat genotypes (Kapulnik and Kushnir 1991). Hetrick et al. (1992) further showed that ancestors and primitive hexaploid wheat landraces benefitted more from mycorrhizal symbiosis than modern cultivars. Subsequently, Hetrick et al. (1993) determined that the ancestral genotype T. tauschii var. strangulata, the donor of the D genotype in hexaploid modern wheat, showed a higher MD as reported in previous studies, whilst AB genome ancestors did not show mycorrhizal dependence. In these studies, the highly fertile conditions used during the plant breeding process were proposed as a possible explanation for the reduced mycorrhizal dependence of modern genotypes. To support this observation, Hetrick et al. (1995) further showed that wheat varieties released before 1975 displayed a higher mycorrhizal responsiveness (MR), defined as the effect of the mycorrhizal symbiosis on plant growth as compared to plants without mycorrhiza, while those released after this date were less responsive. Accordingly, Zhu et al. (2001) also found a reduction in MR in Australian modern wheat cultivars as compared to old cultivars. However, these findings were recently contrasted in a meta-analysis of mycorrhizal responsiveness in wild and annual crop plants. Lehmann et al. (2012) found that newer genotypes were more mycorrhiza-responsive compared to the ancestral genotypes although less intensively colonized. A possible explanation for this observation is that ancient genotypes, and to a larger extent wild relatives, may have developed adaptations to low nutrient environments and are less dependent on mycorrhizal infection than newer genotypes (Koide et al. 1988). However, a decrease in the ability to sustain AMF symbiosis in modern cultivars has been also found for other crops. For instance, it was shown that domesticated breadfruit cultivars (Artocarpus altilis) were less able to support AMF as compared with wild ancestors as revealed by significant reductions of vesicular and arbuscular colonization (Xing et al. 2012). In maize, the response of four landraces and one hybrid to AMF in two different phosphorus (P) regimes was evaluated; two local landraces were significantly more colonized by AMF and acquired more phosphorus in shoots under low and medium P regimes as compared to the modern maize hybrid. Interestingly, one of the landraces presented an outstanding mycorrhizal colonization and presented the highest percentage increase in root volume under both P regimes (Sangabriel-Conde et al. 2014). The diversity of AMF in the roots of the four landraces and the hybrid was assessed through nested PCR of AMF rDNA and it was shown that the landrace with higher mycorrhizal colonization and P acquisition efficiency also presented the highest number of Glomeromycota OTUs (Sangabriel-Conde et al. 2015). The authors proposed that the adoption of native landraces of maize may preserve mycorrhizal symbiosis in these agricultural landscapes.
Domestication effect on rhizobia and other microbes
The effect of plant domestication has also been assessed for the symbiosis between legumes and rhizobia. In a study with pea (Pisum sativum), broad bean (Vicia faba) and several wild legumes from the genera Vicia and Lathyrus, grown in a non-agricultural soil, it was shown that the ability to interact with symbionts was limited for pea and broad bean as compared to promiscuous wild legumes that were able to exploit the diverse rhizobial community (Mutch and Young 2004). Similarly, it was found that Cicer reticulatum, ancestor of cultivated chickpea, showed association with a more diverse Mesorhizobium population than modern chickpea (Kim et al. 2014). In the legume–rhizobia symbiosis, Kiers and Denison (2008) described that plants can sanction less effective symbionts and invest more resources in highly efficient strains. In order to assess whether the ability to sanction non-effective rhizobia strains was also affected in the breeding process, six soybean cultivars representing 60 years of breeding were evaluated in the simultaneous presence of effective and ineffective rhizobia strains. Kiers et al. (2007) showed that newer cultivars had less seed yields as compared to older cultivars and also that the yield difference ratio, i.e. the ability of cultivars to reach their full symbiotic potential in the presence of mixed rhizobial strains, was higher for older cultivars as compared to newer cultivars.
For the effects of plant domestication on other rhizosphere microbes only few examples exist to date. Germida and Siciliano (2001) revealed that the rhizosphere bacterial community of ancient landraces was more diverse than that of two modern cultivars. In the rhizosphere of the ancestral landrace, Pseudomonads were the dominant genus and also higher numbers of Aureobacter were found as compared to modern cultivars (Germida and Siciliano 2001). Also in maize, the influence of its progenitor Zea mays subsp. parviglumis (Balsas teosinte) and two domesticated maize cultivars on the rhizosphere bacterial and fungal community composition was evaluated (Szoboszlay et al. 2015). Shannon’s and Simpson’s diversity indices for bacterial T-RLFP profiles were higher for teosinte compared with one domesticated cultivar and the same as the other cultivar and the control. Interestingly, the same domesticated cultivar with lower bacterial diversity also showed a lower fungal diversity compared with bulk soil controls (Szoboszlay et al. 2015).
Domestication and changes in root architecture
Differences in root architecture between modern cultivars and their wild relatives have been described for a number of crops. For instance, cultivated lettuce (Lactuca sativa) produced a shallower root system compared with wild lettuce (Lactuca serriola). In cultivated lettuce an inadvertent selection of more laterals roots at the top of the tap root helps plants to respond to surface application of water and fertilizer in crop fields, whereas wild lettuce showed a root system able to access deeper portions of soil (Jackson 1995). Changes in root architecture have also been described for drought tolerant plant cultivars. For example, a drought tolerant accession of wild barley presented different root length, root dry weight and root volume compared with a modern cultivar, both under control and drought conditions (Naz et al. 2012, 2014). Similarly, the teosinte Zea mays subsp. parviglumis showed a higher root to shoot dry weight ratio and a higher number of very fine and thick roots than two domesticated maize cultivars. Although the Shannon’s and Simpson’s diversity indices for the bacterial communities were higher for teosinte compared with one the domesticated cultivar, the relative contribution of the root architecture for the observed microbiome differences was not investigated (Szoboszlay et al. 2015). It has been postulated that changes in root architecture due to breeding process may have an effect on the rhizosphere microbiome (Micallef et al. 2009b), however, more detailed studies will be needed to investigate this.
Reinstating beneficial partnerships in modern crop cultivars
Over the past decades, plant breeders have exploited genes from native relatives of modern crop species to improve plant growth and health. For instance, wild relatives have been used as sources of alleles to improve the ability of modern cultivars to withstand biotic and abiotic stresses in wheat (Nevo and Chen 2010; Budak et al. 2013; Placido et al. 2013), barley (Schmalenbach et al. 2008) and lettuce (Johnson et al. 2000; Simko et al. 2013). Similarly, entomologists have explored native habitats to identify natural enemies of insect pests. In the area of plant microbiome research, relatively few efforts have been made to study the biodiversity and functions of beneficial microbial communities present in the native habitats of ancestors of modern crop species. In a study comparing the microbiome of sugar beet and its ancestor Beta vulgaris spp. maritime, plants were grown in agricultural and in native soils (natural habitat). Wild beet plants showed a more diverse bacterial community compared with domesticated sugar beet, as was shown by single strand conformation polymorphism (SSCP) analysis of the 16S-rRNA genes from total community DNA and 16S amplicon pyrosequencing (Zachow et al. 2014). A first approach to identify plant loci associated with root colonization and pathogen protection by beneficial microorganisms involved a study with six inbred tomato lines and the biocontrol bacterium Bacillus cereus UW85 (Smith et al. 1997). Based on a dose–response model, they found differences for both intrinsic plant resistance to pathogen infection and support of biological control in the tomato lines. In a follow-up study with several recombinant inbred line (RIL) populations derived from an interspecific cross of cultivated tomato and the related wild species Lycopersicon cheesmanii, Smith and Goodman (1999) showed that several quantitative trait loci (QTL) were associated with support of growth of the biocontrol agent B. cereus UW85 and the disease suppressive effects. However, several conceptual and experimental efforts have yet to be made in order to identify and exploit these traits in a rhizosphere-based breeding program (Bakker et al. 2012). The search for plant traits linked to microbial recruitment by wild relatives holds a huge potential to elucidate and exploit beneficial interactions between plants and microbes. This hypothesis relies on the assumption that wild plant relatives have coevolved with the microbial community of native soils, performing an active selection of microbes with beneficial effects on plant growth and health. We hypothesize that wild relatives are able to establish, with higher frequency, beneficial interactions with microbes as compared to domesticated cultivars. In this context, we propose a pipeline for this emerging research area (Fig. 3). First, it is necessary to know the evolutionary history and process of domestication of the host plant to make a proper selection of wild plant materials, as well landraces and modern cultivars. If possible, the modern cultivars should be derived from the selected landraces; however this is not possible for all cultivated species, where the full domestication trajectory is unknown. In parallel, the centre of origin and centres of diversification should be known. Botanic and archaeological records have been used to determine where the wild relatives of many modern crops were originally formed as a species, followed by the domestication process and possible routes of dispersion by humans. Using this information, the collection of native soils in pristine sites located in the centre of origin and its use in the experimental setup will provide the native microbial assemblage in which wild relatives presumably recruit and sustain a more beneficial microbiome compared with less competent landraces and modern cultivars. To evaluate the impact of the different plant genotypes on the rhizosphere microbiome composition and functional potential, the use of metagenomics and metatranscriptomics together with culture-dependent approaches can be used to identify shifts in taxonomic and functional diversity of the microbiomes of the different plant genotypes. Based on these “omics” data, a screening with culture-dependent approaches can be performed by targeted isolation of those microbial genera that are specifically or more predominantly recruited by wild plant relatives. Evaluation of antagonistic activities against soil-borne pathogens, nutrient solubilisation or improved drought tolerance of the plant species after introduction will help to pinpoint those beneficial activities that plants look for in microbial partners. Finally, once the recruitment of particular taxa is confirmed and the utility of this association is determined, a plant genotyping strategy, QTL mapping and genome wide association studies (GWAS) with wild relatives, landraces, modern cultivars and preferably crosses between these plant genotypes must be performed in order to identify specific regions in the genome where the recruitment traits are located. Consequently, molecular breeding and marker-assisted selection can be applied to improve beneficial plant–microbe interactions in crop systems. Therefore, this approach of ‘going back to the roots’, i.e. assessing and accessing the microbiome of indigenous plants and their native habitats, represents a yet untapped avenue to further exploit microbes and plant traits in modern agriculture.
References
Badri DV, Vivanco JM (2009) Regulation and function of root exudates. Plant Cell Environ 32(6):666–681. doi:10.1111/j.1365-3040.2008.01926.x
Badri DV, Weir TL, van der Lelie D, Vivanco JM (2009) Rhizosphere chemical dialogues: plant-microbe interactions. Curr Opin Biotechnol 20(6):642–650. doi:10.1016/j.copbio.2009.09.014
Badri DV, Chaparro JM, Zhang R, Shen Q, Vivanco JM (2013) Application of natural blends of phytochemicals derived from the root exudates of Arabidopsis to the soil reveal that phenolic-related compounds predominantly modulate the soil microbiome. J Biol Chem 288(7):4502–4512. doi:10.1074/jbc.M112.433300
Baetz U, Martinoia E (2014) Root exudates: the hidden part of plant defense. Trends Plant Sci 19(2):90–98. doi:10.1016/j.tplants.2013.11.006
Bais HP, Weir TL, Perry LG, Gilroy S, Vivanco JM (2006) The role of root exudates in rhizosphere interactions with plants and other organisms. Annu Rev Plant Biol 57:233–266. doi:10.1146/annurev.arplant.57.032905.105159
Bakker MG, Manter DK, Sheflin AM, Weir TL, Vivanco JM (2012) Harnessing the rhizosphere microbiome through plant breeding and agricultural management. Plant Soil 360(1–2):1–13. doi:10.1007/s11104-012-1361-x
Bakker PAHM, Berendsen RL, Doombos RF, Wintermans PCA, Pieterse CML (2013) The rhizosphere revisited: root microbiomics. Front Plant Sci 4:165. doi:10.3389/fpls.2013.00165
Bednarek P, Kwon C, Schulze-Lefert P (2010) Not a peripheral issue: secretion in plant-microbe interactions. Curr Opin Plant Biol 13(4):378–387. doi:10.1016/j.pbi.2010.05.002
Berendsen RL, Pieterse CMJ, Bakker PAHM (2012) The rhizosphere microbiome and plant health. Trends Plant Sci 17(8):478–486. doi:10.1016/j.tplants.2012.04.001
Berg G, Grube M, Schloter M, Smalla K (2014) Unraveling the plant microbiome: looking back and future perspectives. Front Microbiol 5:148. doi:10.3389/fmicb.2014.00148
Bitocchi E, Nanni L, Bellucci E, Rossi M, Giardini A, Spagnoletti Zeuli P, Logozzo G, Stougaard J, McClean P, Attene G, Papa R (2012) Mesoamerican origin of the common bean (Phaseolus vulgaris L.) is revealed by sequence data. Proc Natl Acad Sci USA 109(14):E788–E796. doi:10.1073/pnas.1108973109
Bitocchi E, Bellucci E, Giardini A, Rau D, Rodriguez M, Biagetti E, Santilocchi R, Spagnoletti Zeuli P, Gioia T, Logozzo G, Attene G, Nanni L, Papa R (2013) Molecular analysis of the parallel domestication of the common bean (Phaseolus vulgaris) in Mesoamerica and the Andes. New Phytol 197:300–313. doi:10.1111/j.1469-8137.2012.04377.x
Bouffaud M-L, Poirier M-A, Mulle D, Moënne-Loccoz Y (2014) Root microbiome relates to plant host evolution in maize and other Poaceae. Environ Microbiol 16:2804–2814. doi:10.1111/1462-2920.12442
Bouwmeester HJ, Roux C, Lopez-Raez JA, Bécard G (2007) Rhizosphere communication of plants, parasitic plants and AM fungi. Trends Plant Sci 12(5):224–230. doi:10.1016/j.tplants.2007.03.009
Broz AK, Manter DK, Vivanco JM (2007) Soil fungal abundance and diversity: another victim of the invasive plant Centaurea maculosa. ISME J 1(8):763–765. doi:10.1038/ismej.2007.81
Budak H, Kantar M, Kurtoglu KY (2013) Drought tolerance in modern and wild wheat. Sci World J. doi:10.1155/2013/548246
Bulgarelli D, Schlaeppi K, Spaepen S, Loren Ver, van Themaat E, Schulze-Lefert P (2013) Structure and functions of the bacterial microbiota of plants. Annu Rev Plant Biol 64:807–838. doi:10.1146/annurev-arplant-050312-120106
Bulgarelli D, Garrido-Oter R, Münch PC, Weiman A, Dröge J, Pan Y, McHardy AC, Schulze-Lefert P (2015) Structure and function of the bacterial root microbiota in wild and domesticated barley. Cell Host Microbe 17(3):392–403. doi:10.1016/j.chom.2015.01.011
Chaparro JM, Badri DV, Vivanco JM (2014) Rhizosphere microbiome assemblage is affected by plant development. ISME J 8:790–803. doi:10.1038/ismej.2013.196
Chen YH, Gols R, Benrey B (2015) Crop domestication and its impact on naturally selected trophic interactions. Annu Rev Entomol 60:35–58. doi:10.1146/annurev-ento-010814-020601
Cook RJ, Thomashow LS, Weller DM, Fujimoto D, Mazzola M, Bangera G, Kim DS (1995) Molecular mechanisms of defense by rhizobacteria against root disease. Proc Natl Acad Sci USA 92(10):4197–4201. doi:10.1073/pnas.92.10.4197
de Weert S, Vermeiren H, Mulders IH, Kuiper I, Hendrickx N, Bloemberg GV, Vanderleyden J, De Mot R, Lugtenberg BJ (2002) Flagella-driven chemotaxis towards exudate components is an important trait for tomato root colonization by Pseudomonas fluorescens. Mol Plant Microbe Interact 15(11):1173–1180. doi:10.1094/MPMI.2002.15.11.1173
de Werra P, Huser A, Tabacchi R, Keel C, Maurhofer M (2011) Plant- and microbe-derived compounds affect the expression of genes encoding antifungal compounds in a pseudomonad with biocontrol activity. Appl Environ Microbiol 77(8):2807–2812. doi:10.1128/AEM.01760-10
Doebley JF, Gaut BS, Smith BD (2006) The molecular genetics of crop domestication. Cell 127(7):1309–1321. doi:10.1016/j.cell.2006.12.006
Fierer N, Ladau J, Clemente JC, Leff JW, Owens SM, Pollard KS, Knight R, Gilbert JA, McCulley RL (2013) Reconstructing the microbial diversity and function of pre-agricultural tallgrass prairie soils in the United States. Science 342(6158):621–624. doi:10.1126/science.1243768
García-Palacios P, Milla R, Delgado-Baquerizo M, Martín-Robles N, Alvaro-Sánchez M, Wall DH (2013) Side-effects of plant domestication: ecosystem impacts of changes in litter quality. New Phytol 198(2):504–513. doi:10.1111/nph.12127
Gepts P (2004) Crop domestication as a long-term selection experiment. Plant Breed Rev 24(2):1–44. doi:10.1002/9780470650288.ch1
Germida JJ, Siciliano SD (2001) Taxonomic diversity of bacteria associated with the roots of modern, recent and ancient wheat cultivars. Biol Fertil Soils 33:410–415. doi:10.1007/s003740100343
Gomes NCM, Heuer H, Schonfeld J, Costa R, Mendonca-Hagler L, Smalla K (2001) Bacterial diversity of the rhizosphere of maize (Zea mays) grown in tropical soil studied by temperature gradient gel electrophoresis. Plant Soil 232:167–180. doi:10.1023/A:1010350406708
Gross BL, Olsen KM (2010) Genetic perspectives on crop domestication. Trends Plant Sci 15(9):529–537. doi:10.1016/j.tplants.2010.05.008
Haichar FZ, Marol C, Berge O, Rangel-Castro JI, Prosser JI, Balesdent J, Heulin T, Achouak W (2008) Plant host habitat and root exudates shape soil bacterial community structure. ISME J 2(12):1221–1230. doi:10.1038/ismej.2008.80
Haichar FZ, Santaella C, Heulin T, Achouak W (2014) Root exudates mediated interactions belowground. Soil Biol Biochem 77:69–80. doi:10.1016/j.soilbio.2014.06.017
Haudry A, Cenci A, Ravel C, Bataillon T, Brunel D, Poncet C, Hochu I, Poirier S, Santoni S, Glémin S, David J (2007) Grinding up wheat: a massive loss of nucleotide diversity since domestication. Mol Biol Evol 24(7):1506–1517. doi:10.1093/molbev/msm077
Henry S, Texier S, Hallet S, Bru D, Dambreville C, Chèneby D, Bizouard F, Germon JC, Philippot L (2008) Disentangling the rhizosphere effect on nitrate reducers and denitrifiers: insight into the role of root exudates. Environ Microbiol 10(11):3082–3092. doi:10.1111/j.1462-2920.2008.01599.x
Hetrick BAD, Wilson GWT, Cox TS (1992) Mycorrhizal dependence of modern wheat varieties, landraces, and ancestors. Can J Bot 70(10):2032–2040. doi:10.1139/b92-253
Hetrick BAD, Wilson GWT, Cox TS (1993) Mycorrhizal dependence of modern wheat cultivars and ancestors: a synthesis. Can J Bot 71(3):512–518. doi:10.1139/b93-056
Hetrick BAD, Wilson GWT, Gill BS, Cox TS (1995) Chromosome location of mycorrhizal responsive genes in wheat. Can J Bot 73:891–897. doi:10.1139/b95-097
Inceoglu Ö, Falcão Salles J, van Overbeek L, van Elsas JD (2010) Effects of plant genotype and growth stage on the betaproteobacterial communities associated with different potato cultivars in two fields. Appl Environ Microbiol 76(11):3675–3684. doi:10.1128/AEM.00040-10
Inceoglu Ö, Abu Al-Soud W, Salles JF, Semenov AV, van Elsas JD (2011) Comparative analysis of bacterial communities in a potato field as determined by pyrosequencing. PLoS One 6(8):e23321. doi:10.1371/journal.pone.0023321
Jackson LE (1995) Root architecture in cultivated and wild lettuce (Lactuca app). Plant Cell Environ 18:885–894. doi:10.1111/j.1365-3040.1995.tb00597.x
Jin J, Wang GH, Liu XB, Liu JD, Chen XL, Herbert SJ (2009) Temporal and spatial dynamics of bacterial community in the rhizosphere of soybean genotypes grown in a black soil. Pedosphere 19(6):808–816. doi:10.1016/S1002-0160(09)60176-4
Johnson WC, Jackson LE, Ochoa O, van Wijk R, Peleman J, Clair DA, Michelmore RW (2000) Lettuce, a shallow-rooted crop, and Lactuca serriola, its wild progenitor, differ at QTL determining root architecture and deep soil water exploitation. Theor Appl Genet 101:1066–1073. doi:10.1007/s001220051581
Jones DL, Hodge A, Kuzyakov Y (2004) Plant and mycorrhizal regulation of rhizodeposition. New Phytol 163:459–480. doi:10.1111/j.1469-8137.2004.01130.x
Jones DL, Nguyen C, Finlay RD (2009) Carbon flow in the rhizosphere: carbon trading at the soil–root interface. Plant Soil 321:5–33. doi:10.1007/s11104-009-9925-0
Jousset A, Rochat L, Lanoue A, Bonkowski M, Keel C, Scheu S (2011) Plants respond to pathogen infection by enhancing the antifungal gene expression of root-associated bacteria. Mol Plant Microbe Interact 24(3):352–358. doi:10.1094/MPMI-09-10-0208
Kapulnik Y, Kushnir U (1991) Growth dependency of wild, primitive and modern cultivated wheat lines on vesicular–arbuscular mycorrhiza fungi. Euphytica 56:27–36. doi:10.1007/BF00041740
Kiers ET, Denison RF (2008) Sanctions, cooperation, and the stability of plant-rhizosphere mutualisms. Annu Rev Ecol Evol Syst 39:215–236. doi:10.1146/annurev.ecolsys.39.110707.173423
Kiers ET, Hutton MG, Denison RF (2007) Human selection and the relaxation of legume defences against ineffective rhizobia. Proc Biol Sci 274(1629):3119–3126. doi:10.1098/rspb.2007.1187
Kim DH, Kaashyap M, Rathore A, Das RR, Parupalli S, Upadhyaya HD, Gopalakrishnan S, Gaur PM, Singh S, Kaur J, Yasin M, Varshney RK (2014) Phylogenetic diversity of Mesorhizobium in chickpea. J Biosci 39(3):513–517. doi:10.1007/s12038-014-9429-9
Koide R, Li M, Lewis J, Irby C (1988) Role of mycorrhizal infection in the growth and reproduction of wild vs. cultivated plants. I. wild vs. cultivated oats. Oecologia 77:537–543. doi:10.1007/BF00377271
Köllner TG, Held M, Lenk C, Hiltpold I, Turlings TC, Gershenzon J, Degenhardt J (2008) A maize (E)-beta-caryophyllene synthase implicated in indirect defense responses against herbivores is not expressed in most American maize varieties. Plant Cell 20(2):482–494. doi:10.1105/tpc.107.051672
Lakshmanan V, Kitto SL, Caplan JL, Hsueh YH, Kearns DB, Wu YS, Bais HP (2012) Microbe-associated molecular patterns-triggered root responses mediate beneficial rhizobacterial recruitment in Arabidopsis. Plant Physiol 160(3):1642–1661. doi:10.1104/pp.112.200386
Lakshmanan V, Selvaraj G, Bais HP (2014) Functional soil microbiome: belowground solutions to an aboveground problem. Plant Physiol 166(2):689–700. doi:10.1104/pp.114.245811
Lehmann A, Barto EK, Powell JR, Rillig MC (2012) Mycorrhizal responsiveness trends in annual crop plants and their wild relatives—a meta-analysis on studies from 1981 to 2010. Plant Soil 355:231–250. doi:10.1007/s11104-011-1095-1
Ling N, Raza W, Ma J, Huang Q, Shen Q (2011) Identification and role of organic acids in watermelon root exudates for recruiting Paenibacillus polymyxa SQR-21 in the rhizosphere. Eur J Soil Biol 47:374–379. doi:10.1016/j.ejsobi.2011.08.009
Marques JM, da Silva TF, Vollu RE, Blank AF, Ding GC, Seldin L, Smalla K (2014) Plant age and genotype affect the bacterial community composition in the tuber rhizosphere of field-grown sweet potato plants. FEMS Microbiol Ecol 88(2):424–435. doi:10.1111/1574-6941.12313
Matson PA, Parton WJ, Power AG, Swift MJ (1997) Agricultural intensification and ecosystem properties. Science 277(5325):504–509. doi:10.1126/science.277.5325.504
Mendes R, Kruijt M, de Bruijn I, Dekkers E, van der Voort M, Schneider JHM, Piceno Y, DeSantis TZ, Andersen GL, Bakker PAHM, Raaijmakers JM (2011) Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 332(6033):1097–1100. doi:10.1126/science.1203980
Mendes R, Garbeva P, Raaijmakers JM (2013) The rhizosphere microbiome: significance of plant beneficial, plant pathogenic and human pathogenic microorganisms. FEMS Microbiol Rev 37:634–663. doi:10.1111/1574-6976.12028
Micallef SA, Channer S, Shiaris MP, Colón-Carmona A (2009a) Plant age and genotype impact the progression of bacterial community succession in the Arabidopsis rhizosphere. Plant Signal Behav 4(8):777–780. doi:10.1093/jxb/erp053
Micallef SA, Shiaris MP, Colón-Carmona A (2009b) Influence of Arabidopsis thaliana accessions on rhizobacterial communities and natural variation in root exudates. J Exp Bot 60(6):1729–1742. doi:10.1093/jxb/erp053
Moe LA (2013) Amino acids in the rhizosphere: from plants to microbes. Am J Bot 100(9):1692–1705. doi:10.3732/ajb.1300033
Mounier E, Hallet S, Chèneby D, Benizri E, Gruet Y, Nguyen C, Piutti S, Robin C, Slezack-Deschaumes S, Martin-Laurent F, Germon JC, Philippot L (2004) Influence of maize mucilage on the diversity and activity of the denitrifying community. Environ Microbiol 6(3):301–312. doi:10.1111/j.1462-2920.2004.00571.x
Mummey DL, Rillig MC (2006) The invasive plant species Centaurea maculosa alters arbuscular mycorrhizal fungal communities in the field. Plant Soil 288:81–90. doi:10.1007/s11104-006-9091-6
Mutch LA, Young JP (2004) Diversity and specificity of Rhizobium leguminosarum biovar viciae on wild and cultivated legumes. Mol Ecol 13(8):2435–2444. doi:10.1111/j.1365-294X.2004.02259.x
Naz AA, Ehl A, Pillen K, Léon J (2012) Validation for root-related quantitative trait locus effects of wild origin in the cultivated background of barley (Hordeum vulgare L.). Plant Breed 3:392–398. doi:10.1111/j.1439-0523.2012.01972.x
Naz AA, Arifuzzaman M, Muzammil S, Pillen K, Léon J (2014) Wild barley introgression lines revealed novel QTL alleles for root and related shoot traits in the cultivated barley (Hordeum vulgare L.). BMC Genet 15:107. doi:10.1186/s12863-014-0107-6
Neal AL, Ahmad S, Gordon-Weeks R, Ton J (2012) Benzoxazinoids in root exudates of maize attract Pseudomonas putida to the rhizosphere. PLoS One 7(4):e35498. doi:10.1371/journal.pone.0035498
Nevo E, Chen G (2010) Drought and salt tolerances in wild relatives for wheat and barley improvement. Plant Cell Environ 33:670–685. doi:10.1111/j.1365-3040.2009.02107.x
Ofek M, Voronov-Goldman M, Hadar Y, Minz D (2014) Host signature effect on plant root-associated microbiomes revealed through analyses of resident vs. active communities. Environ Microbiol 16:2157–2167. doi:10.1111/1462-2920.12228
Olsen KM, Caicedo AL, Polato N, McClung A, McCouch S, Purugganan MD (2006) Selection under domestication: evidence for a sweep in the rice waxy genomic region. Genetics 173(2):975–983. doi:10.1534/genetics.106.056473
Palaisa K, Morgante M, Tingey S, Rafalski A (2004) Long-range patterns of diversity and linkage disequilibrium surrounding the maize Y1 gene are indicative of an asymmetric selective sweep. Proc Natl Acad Sci USA 101(26):9885–9890. doi:10.1073/pnas.0307839101
Peiffer JA, Spor A, Koren O, Jin Z, Tringe SG, Dangl JL, Buckler ES, Ley RE (2013) Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc Natl Acad Sci USA 110(16):6548–6553. doi:10.1073/pnas.1302837110
Placido DF, Campbell MT, Folsom JJ, Cui X, Kruger GR, Baenziger PS, Harkamal WH (2013) Introgression of novel traits from a wild wheat relative improves drought adaptation in wheat. Plant Physiol 161(4):1806–1819. doi:10.1104/pp.113.214262
Pongsilp N, Nimnoi P, Lumyong S (2012) Genotypic diversity among rhizospheric bacteria of three legumes assessed by cultivation-dependent and cultivation-independent techniques. World J Microbiol Biotechnol 28(2):615–626. doi:10.1007/s11274-011-0855-7
Purugganan MD, Fuller DQ (2009) The nature of selection during plant domestication. Nature 457:843–848. doi:10.1038/nature07895
Ram SG, Thiruvengadam V, Vinod KK (2007) Genetic diversity among cultivars, landraces and wild relatives of rice as revealed by microsatellite markers. J Appl Genet 48(4):337–345. doi:10.1007/BF03195230
Ramirez KS, Craine JM, Fierer N (2012) Consistent effects of nitrogen amendments on soil microbial communities and processes across biomes. Glob Change Biol 18:1918–1927. doi:10.1111/j.1365-2486.2012.02639.x
Rasmann S, Köllner TG, Degenhardt J, Hiltpold I, Toepfer S, Kuhlmann U, Gershenzon J, Turlings TC (2005) Recruitment of entomopathogenic nematodes by insect-damaged maize roots. Nature 434(7034):732–737. doi:10.1038/nature03451
Rodrigues JL, Pellizari VH, Mueller R, Baek K, Jesus Eda C, Paula FS, Mirza B, Hamaoui GS Jr, Tsai SM, Feigl B, Tiedje JM, Bohannan BJ, Nüsslein K (2013) Conversion of the Amazon rainforest to agriculture results in biotic homogenization of soil bacterial communities. Proc Natl Acad Sci USA 110(3):988–993. doi:10.1073/pnas.1220608110
Rudrappa T, Czymmek KJ, Paré PW, Bais HP (2008) Root-secreted malic acid recruits beneficial soil bacteria. Plant Physiol 148(3):1547–1556. doi:10.1104/pp.108.127613
Sangabriel-Conde W, Negrete-Yankelevich S, Maldonado-Mendoza IE, Trejo-Aguilar D (2014) Native maize landraces from Los Tuxtlas, Mexico show varying mycorrhizal dependency for P uptake. Biol Fertil Soils 50:405–414. doi:10.1007/s00374-013-0847-x
Sangabriel-Conde W, Maldonado-Mendoza IE, Mancera-López ME, Cordero-Ramírez JD, Trejo-Aguilar D, Negrete-Yankelevich S (2015) Glomeromycota associated with Mexican native maize landraces in Los Tuxtlas, Mexico. Appl Soil Ecol 87:63–71. doi:10.1016/j.apsoil.2014.10.017
Schmalenbach I, Körber N, Pillen K (2008) Selecting a set of wild barley introgression lines and verification of QTL effects for resistance to powdery mildew and leaf rust. Theor Appl Genet 117(7):1093–1106. doi:10.1007/s00122-008-0847-7
Shi S, Richardson AE, O’Callaghan M, DeAngelis KM, Jones EE, Stewart A, Firestone MK, Condron LM (2011) Effects of selected root exudate components on soil bacterial communities. FEMS Microbiol Ecol 77(3):600–610. doi:10.1111/j.1574-6941.2011.01150.x
Simko I, Atallah AJ, Ochoa OE, Antonise R, Galeano CH, Truco MJ, Michelmore MW (2013) Identification of QTLs conferring resistance to downy mildew in legacy cultivars of lettuce. Sci Rep 3:2875. doi:10.1038/srep02875
Smalla K, Wieland G, Buchner A, Zock A, Parzy J, Kaiser S, Roskot N, Heuer H, Berg G (2001) Bulk and rhizosphere soil bacterial communities studied by denaturing gradient gel electrophoresis: plant-dependent enrichment and seasonal shifts revealed. Appl Environ Microbiol 67(10):4742–4751. doi:10.1128/AEM.67.10.4742-4751.2001
Smith KP, Goodman RM (1999) Host variation for interactions with beneficial plant-associated microbes. Annu Rev Phytopathol 37:473–491. doi:10.1146/annurev.phyto.37.1.473
Smith KP, Handelsman J, Goodman RM (1997) Modeling dose-response relationships in biological control: partitioning host responses to the pathogen and biocontrol agent. Phytopathol 87(7):720–729. doi:10.1094/PHYTO.1997.87.7.720
Sugiyama A, Ueda Y, Takase H, Yazaki K (2014a) Pyrosequencing assessment of rhizosphere fungal communities from a soybean field. Can J Microbiol 60(10):687–690. doi:10.1139/cjm-2014-0443
Sugiyama A, Ueda Y, Zushi T, Takase H, Yazaki K (2014b) Changes in the bacterial community of soybean rhizospheres during growth in the field. PLoS One 9(6):e100709. doi:10.1371/journal.pone.0100709
Szoboszlay M, Lambers J, Chappell J, Kupper JV, Moe LA, McNear DH Jr (2015) Comparison of root system architecture and rhizosphere microbial communities of Balsas teosinte and domesticated corn cultivars. Soil Biol Biochem 80:34–44. doi:10.1016/j.soilbio.2014.09.001
Turner TR, James EK, Poole PS (2013a) The plant microbiome. Genome Biol 14:209. doi:10.1186/gb-2013-14-6-209
Turner TR, Ramakrishnan K, Walshaw J, Heavens D, Alston M, Swarbreck D, Osbourn A, Grant A, Poole PS (2013b) Comparative metatranscriptomics reveals kingdom level changes in the rhizosphere microbiome of plants. ISME J 7(12):2248–2258. doi:10.1038/ismej.2013.119
Wang E, Schornack S, Marsh JF, Gobbato E, Schwessinger B, Eastmond P, Schultze M, Kamoun S, Oldroyd GE (2012) A common signaling process that promotes mycorrhizal and oomycete colonization of plants. Curr Biol 22(23):2242–2246. doi:10.1016/j.cub.2012.09.043
Weese DJ, Heath KD, Dentinger BT, Lau JA (2015) Long-term nitrogen addition causes the evolution of less-cooperative mutualists. Evolution 69(3):631–642. doi:10.1111/evo.12594
Weinert N, Piceno Y, Ding GC, Meincke R, Heuer H, Berg G, Schloter M, Andersen G, Smalla K (2011) PhyloChip hybridization uncovered an enormous bacterial diversity in the rhizosphere of different potato cultivars: many common and few cultivar-dependent taxa. FEMS Microbiol Ecol 75(3):497–506. doi:10.1111/j.1574-6941.2010.01025.x
Weston LA, Mathesius U (2013) Flavonoids: their structure, biosynthesis and role in the rhizosphere, including allelopathy. J Chem Ecol 39(2):283–297. doi:10.1007/s10886-013-0248-5
Wieland G, Neumann R, Backhaus H (2001) Variation of microbial communities in soil, rhizosphere, and rhizoplane in response to crop species, soil type, and crop development. Appl Environ Microbiol 67(12):5849–5854
Wissuwa M, Mazzola M, Picard C (2009) Novel approaches in plant breeding for rhizosphere related traits. Plant Soil 321:409–430. doi:10.1007/s11104-008-9693-2
Xing X, Koch AM, Jones AM, Ragone D, Murch S, Hart MM (2012) Mutualism breakdown in breadfruit domestication. Proc Biol Sci 22(279(1731)):1122–1130. doi:10.1098/rspb.2011.1550
Xu Y, Wang G, Jin J, Liu J, Zhang Q, Liu X (2009) Bacterial communities in soybean rhizosphere in response to soil type, soybean genotype, and their growth stage. Soil Biol Biochem 41:919–925. doi:10.1016/j.soilbio.2008.10.027
Zachow C, Müller H, Tilcher R, Berg G (2014) Differences between the rhizosphere microbiome of Beta vulgaris ssp. maritima—ancestor of all beet crops—and modern sugar beets. Front Microbiol 5:415. doi:10.3389/fmicb.2014.00415
Zhang N, Wang D, Liu Y, Li S, Shen Q, Zhang R (2014) Effects of different plant root exudates and their organic acid components on chemotaxis, biofilm formation and colonization by beneficial rhizosphere-associated bacterial strains. Plant Soil 374:689–700. doi:10.1007/s11104-013-1915-6
Zhu Y-G, Smith SE, Barritt AR, Smith FA (2001) Phosphorus (P) efficiencies and mycorrhizal responsiveness of old and modern wheat cultivars. Plant Soil 237:249–255. doi:10.1023/A:1013343811110
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Pérez-Jaramillo, J.E., Mendes, R. & Raaijmakers, J.M. Impact of plant domestication on rhizosphere microbiome assembly and functions. Plant Mol Biol 90, 635–644 (2016). https://doi.org/10.1007/s11103-015-0337-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-015-0337-7