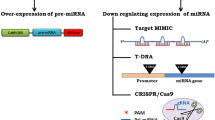
Abstract
Food security is one of the most important issues challenging the world today. Any strategies to solve this problem must include increasing crop yields and quality. MicroRNA-based genetic modification technology (miRNA-based GM tech) can be one of the most promising solutions that contribute to agricultural productivity directly by developing superior crop cultivars with enhanced biotic and abiotic stress tolerance and increased biomass yields. Indirectly, the technology may increase usage of marginal soils and decrease pesticide use, among other benefits. This review highlights the most recent progress of transgenic studies utilizing various miRNAs and their targets for plant trait modifications, and analyzes the potential of miRNA-mediated gene regulation for use in crop improvement. Strategies for manipulating miRNAs and their targets in transgenic plants including constitutive, stress-induced, or tissue-specific expression of miRNAs or their targets, RNA interference, expressing miRNA-resistant target genes, artificial target mimic and artificial miRNAs were discussed. We also discussed potential risks of utilizing miRNA-based GM tech. In general, miRNAs and their targets not only provide an invaluable source of novel transgenes, but also inspire the development of several new GM strategies, allowing advances in breeding novel crop cultivars with agronomically useful characteristics.


Similar content being viewed by others
References
Allen RS, Millgate AG, Chitty JA, Thisleton J, Miller JA, Fist AJ, Gerlach WL, Larkin PJ (2004) RNAi-mediated replacement of morphine with the nonnarcotic alkaloid reticuline in opium poppy. Nat Biotechnol 22:1559–1566
Alvarez JP, Pekker I, Goldshmidt A, Blum E, Amsellem Z, Eshed Y (2006) Endogenous and synthetic microRNAs stimulate simultaneous, efficient, and localized regulation of multiple targets in diverse species. Plant Cell 18:1134–1151
Aukerman MJ, Sakai H (2003) Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 15:2730–2741
Axtell MJ, Bowman JL (2008) Evolution of plant microRNAs and their targets. Trends Plant Sci 13:343–349
Baum JA, Bogaert T, Clinton W, Heck GR, Feldmann P, Ilagan O, Johnson S, Plaetinck G, Munyikwa T, Pleau M (2007) Control of coleopteran insect pests through RNA interference. Nat Biotechnol 25:1322–1326
Bazzini A, Hopp H, Beachy R, Asurmendi S (2007) Infection and coaccumulation of tobacco mosaic virus proteins alter microRNA levels, correlating with symptom and plant development. Proc Natl Acad Sci USA 104:12157–12162
Beddington J (2010) Food security: contributions from science to a new and greener revolution. Phil Trans R Soc B 365:61–71
Bowman JL, Smyth DR, Meyerowitz EM (1991) Genetic interactions among floral homeotic genes of Arabidopsis. Development 112:1–20
Bowman JL, Smyth DR, Meyerowitz EM (2012) The ABC model of flower development: then and now. Development 139:4095–4098
Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, Voinnet O (2008) Widespread translational inhibition by plant miRNAs and siRNAs. Science 320:1185–1190
Buiatti M, Christou P, Pastore G (2012) The application of GMOs in agriculture and in food production for a better nutrition: two different scientific points of view. Genes Nutr 8:255–270
Byrnes BH, Bumb BL (1998) Population growth, food production and nutrient requirements. J Crop Prod 1:1–27
Campo S, Peris-Peris C, Siré C, Moreno AB, Donaire L, Zytnicki M, Notredame C, Llave C, San Segundo B (2013) Identification of a novel microRNA (miRNA) from rice that targets an alternatively spliced transcript of the Nramp6 (Natural resistance-associated macrophage protein 6) gene involved in pathogen resistance. New Phytol. doi:10.1111/nph.12292
Capote N, Pérez-Panadés J, Monzó C, Carbonell E, Urbaneja A, Scorza R, Ravelonandro M, Cambra M (2008) Assessment of the diversity and dynamics of Plum pox virus and aphid populations in transgenic European plums under Mediterranean conditions. Transgenic Res 17:367–377
Century K, Reuber TL, Ratcliffe OJ (2008) Regulating the regulators: the future prospects for transcription-factor-based agricultural biotechnology products. Plant Physiol 147:20–29
Chen L, Wang T, Zhao M, Tian Q, Zhang WH (2012) Identification of aluminum-responsive microRNAs in Medicago truncatula by genome-wide high-throughput sequencing. Planta 235:375–386
Christou P, Twyman RM (2004) The potential of genetically enhanced plants to address food insecurity. Nutr Res Rev 17:23–42
Chuck GS, Tobias C, Sun L, Kraemer F, Li C, Dibble D, Arora R, Bragg JN, Vogel JP, Singh S (2011) Overexpression of the maize Corngrass1 microRNA prevents flowering, improves digestibility, and increases starch content of switchgrass. Proc Natl Acad Sci USA 108:17550–17555
Coca MA, Almoguera C, Thomas TL, Jordano J (1996) Differential regulation of small heat-shock genes in plants: analysis of a water-stress-inducible and developmentally activated sunflower promoter. Plant Mol Biol 31:863–876
Comai L, Zhang B (2012) MicroRNAs: key gene regulators with versatile functions. Plant Mol Biol. doi:10.1007/s11103-012-9947-5
Cuperus JT, Fahlgren N, Carrington JC (2011) Evolution and functional diversification of MIRNA genes. Plant Cell 23:431–442
Datta K, Vasquez A, Tu J, Torrizo L, Alam M, Oliva N, Abrigo E, Khush G, Datta S (1998) Constitutive and tissue-specific differential expression of the cryIA (b) gene in transgenic rice plants conferring resistance to rice insect pest. Theor Appl Genet 97:20–30
Davuluri GR, Van Tuinen A, Fraser PD, Manfredonia A, Newman R, Burgess D, Brummell DA, King SR, Palys J, Uhlig J (2005) Fruit-specific RNAi-mediated suppression of DET1 enhances carotenoid and flavonoid content in tomatoes. Nat Biotechnol 23:890–895
Ding YF, Zhu C (2009) The role of microRNAs in copper and cadmium homeostasis. Biochem Biophys Res Commun 386:6–10
Ding Y, Chen Z, Zhu C (2011) Microarray-based analysis of cadmium-responsive microRNAs in rice (Oryza sativa). J Exp Bot 62:3563–3573
Dunoyer P, Voinnet O (2009) Movement of RNA silencing between plant cells: is the question now behind us? Trends Plant Sci 14:643–644
Eckardt NA (2012) A microRNA cascade in plant defense. Plant Cell 24:840
Fahlgren N, Howell MD, Kasschau KD, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Law TF, Grant SR, Dangl JL, Carrington JC (2007) High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA Genes. PLoS ONE 2:e219. doi:10.1371/journal.pone.0000219
FAO (1996) Declaration on world food security. World Food Summit, FAO, Rome. http://www.fao.org/docrep/003/w3613e/w3613e00.htm. Accessed 28 Jan 2013
FAO (2008) Briefing paper: hunger on the rise. FAO, Rome, Italy. http://www.fao.org/newsroom/common/ecg/1000923/en/hungerfigs.pdf. Accessed 5 Feb 2013
FAO Statistics Division (2006) Food deprivation trends: midterm review of progress towards the World Food Summit target. Working paper series WP007e, FAO, Rome, Italy. http://www.fao.org/docrep/013/am064e/am064e00.pdf. Accessed 5 Feb 2013
Farre G, Twyman RM, Zhu C, Capell T, Christou P (2011) Nutritionally enhanced crops and food security: scientific achievements versus political expediency. Curr Opin Biotechnol 22:245–251
Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, Garcia JA, Paz-Ares J (2007) Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet 39:1033–1037
Fu C, Sunkar R, Zhou C, Shen H, Zhang J-Y, Matts J, Wolf J, Mann DGJ, Stewart CN, Tang Y, Wang ZY (2012) Overexpression of miR156 in switchgrass (Panicum virgatum L.) results in various morphological alterations and leads to improved biomass production. Plant Biotechnol J 10:443–452
Fuchs M, Gonsalves D (2007) Safety of virus-resistant transgenic plants two decades after their introduction: lessons from realistic field risk assessment studies. Annu Rev Phytopathol 45:173–202
Fujii H, Chiou T-J, Lin S-I, Aung K, Zhu J-K (2005) A miRNA involved in phosphate-starvation response in Arabidopsis. Curr Biol 15:2038–2043
Fujii N, Inui T, Iwasa K, Morishige T, Sato F (2007) Knockdown of berberine bridge enzyme by RNAi accumulates (S)-reticuline and activates a silent pathway in cultured California poppy cells. Transgenic Res 16:363–375
Gao P, Bai X, Yang L, Lv D, Li Y, Cai H, Ji W, Guo D, Zhu Y (2010) Over-expression of osa-MIR396c decreases salt and alkali stress tolerance. Planta 231:991–1001. doi:10.1007/s00425-010-1104-2
Gao P, Bai X, Yang L, Lv D, Pan X, Li Y, Cai H, Ji W, Chen Q, Zhu Y (2011) osa-MIR393: a salinity- and alkaline stress-related microRNA gene. Mol Biol Rep 38:237–242. doi:10.1007/s11033-010-0100-8
Garg AK, Kim JK, Owens TG, Ranwala AP, Do Choi Y, Kochian LV, Wu RJ (2002) Trehalose accumulation in rice plants confers high tolerance levels to different abiotic stresses. Proc Natl Acad Sci USA 99:15898–15903
Godfray HCJ, Beddington JR, Crute IR, Haddad L, Lawrence D, Muir JF, Pretty J, Robinson S, Thomas SM, Toulmin C (2010) Food security: the challenge of feeding 9 billion people. Science 327:812
Goetz M, Hooper LC, Johnson SD, Rodrigues JCM, Vivian-Smith A, Koltunow AM (2007) Expression of aberrant forms of AUXIN RESPONSE FACTOR8 stimulates parthenocarpy in Arabidopsis and tomato. Plant Physiol 145:351–366
Guan Q, Lu X, Zeng H, Zhang Y, Zhu J (2013) Heat stress induction of miR398 triggers a regulatory loop that is critical for thermotolerance in Arabidopsis. Plant J. doi:10.1111/tpj.12169
Guo S, Xu Y, Liu H, Mao Z, Zhang C, Ma Y, Zhang Q, Meng Z, Chong K (2013) The interaction between OsMADS57 and OsTB1 modulates rice tillering via DWARF14. Nat Commun. doi:10.1038/ncomms2542
Gupta AS, Heinen JL, Holaday AS, Burke JJ, Allen RD (1993) Increased resistance to oxidative stress in transgenic plants that overexpress chloroplastic Cu/Zn superoxide dismutase. Proc Natl Acad Sci USA 90:1629–1633
He XF, Fang YY, Feng L, Guo HS (2008) Characterization of conserved and novel microRNAs and their targets, including a TuMV-induced TIR–NBS–LRR class R gene-derived novel miRNA in Brassica. FEBS Lett 582:2445–2452
Hewezi T, Howe P, Maier TR, Baum TJ (2008) Arabidopsis small RNAs and their targets during cyst nematode parasitism. Mol Plant Microbe Interact 21:1622–1634
Hsieh YT, Pan TM (2006) Influence of planting papaya ringspot virus resistant transgenic papaya on soil microbial biodiversity. J Agric Food Chem 54:130–137
Huang G, Allen R, Davis EL, Baum TJ, Hussey RS (2006) Engineering broad root-knot resistance in transgenic plants by RNAi silencing of a conserved and essential root-knot nematode parasitism gene. Proc Natl Acad Sci USA 103:14302–14306
Huang SQ, Peng J, Qiu CX, Yang ZM (2009) Heavy metal-regulated new microRNAs from rice. J Inorg Biochem 103:282–287
Huang SQ, Xiang AL, Che LL, Chen S, Li H, Song JB, Yang ZM (2010) A set of miRNAs from Brassica napus in response to sulphate deficiency and cadmium stress. Plant Biotechnol J 8:887–899
IMF (2008) IMF primary commodity prices. IMF, Washington, DC. http://www.imf.org/external/np/res/commod/. Accessed 5 Feb 2013
Itaya A, Bundschuh R, Archual AJ, Joung JG, Fei Z, Dai X, Zhao PX, Tang Y, Nelson RS, Ding B (2008) Small RNAs in tomato fruit and leaf development. Biochim Biophys Acta 1779:99–107
Ivashuta SI, Petrick JS, Heisel SE, Zhang Y, Guo L, Reynolds TL, Rice JF, Allen E, Roberts JK (2009) Endogenous small RNAs in grain: semi-quantification and sequence homology to human and animal genes. Food Chem Toxicol 47:353–360
Jagadeeswaran G, Saini A, Sunkar R (2009) Biotic and abiotic stress down-regulate miR398 expression in Arabidopsis. Planta 229:1009–1014
James C (2011) Global status of commercialized biotech/GM crops: 2011. ISAAA brief 43; ISAAA: Ithaca, NY. http://wwwisaaaorg/resources/publications/briefs/43/defaultasp. Accessed 5 Feb 2013
Jiao Y, Wang Y, Xue D, Wang J, Yan M, Liu G, Dong G, Zeng D, Lu Z, Zhu X (2010) Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat Genet 42:541–544
Jones-Rhoades MW (2011) Conservation and divergence in plant microRNAs. Plant Mol Biol 80:3–16
Jones-Rhoades MW, Bartel DP, Bartel B (2006) MicroRNAs and their regulatory roles in plants. Annu Rev Plant Biol 57:19–53
Karlova R, van Haarst JC, Maliepaard C, van de Geest H, Bovy AG, Lammers M, Angenent GC, de Maagd RA (2013) Identification of microRNA targets in tomato fruit development using high-throughput sequencing and degradome analysis. J Exp Bot. doi:10.1093/jxb/ert049
Kasschau KD, Xie Z, Allen E, Llave C, Chapman EJ, Krizan KA, Carrington JC (2003) P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA function. Dev Cell 4:205–217
Kasuga M, Miura S, Shinozaki K, Yamaguchi-Shinozaki K (2004) A combination of the Arabidopsis DREB1A gene and stress-inducible rd29A promoter improved drought-and low-temperature stress tolerance in tobacco by gene transfer. Plant Cell Physiol 45:346–350
Katiyar-Agarwal S, Jin H (2010) Role of small RNAs in host-microbe interactions. Annu Rev Phytopathol 48:225–246
Katiyar-Agarwal S, Morgan R, Dahlbeck D, Borsani O, Villegas A Jr, Zhu JK, Staskawicz BJ, Jin H (2006) A pathogen-inducible endogenous siRNA in plant immunity. Proc Natl Acad Sci USA 103:18002–18007
Katiyar-Agarwal S, Gao S, Vivian-Smith A, Jin H (2007) A novel class of bacteria-induced small RNAs in Arabidopsis. Genes Dev 21:3123–3134
Kawashima CG, Yoshimoto N, Maruyama-Nakashita A, Tsuchiya YN, Saito K, Takahashi H, Dalmay T (2008) Sulphur starvation induces the expression of microRNA-395 and one of its target genes but in different cell types. Plant J 57:313–321
Kawashima CG, Matthewman CA, Huang S, Lee BR, Yoshimoto N, Koprivova A, Rubio-Somoza I, Todesco M, Rathjen T, Saito K (2011) Interplay of SLIM1 and miR395 in the regulation of sulfate assimilation in Arabidopsis. Plant J 66:863–876
Khraiwesh B, Ossowski S, Weigel D, Reski R, Frank W (2008) Specific gene silencing by artificial MicroRNAs in Physcomitrella patens: an alternative to targeted gene knockouts. Plant Physiol 148:684–693
Khraiwesh B, Arif MA, Seumel GI, Ossowski S, Weigel D, Reski R, Frank W (2010) Transcriptional control of gene expression by microRNAs. Cell 140:111–122
Khraiwesh B, Zhu J-K, Zhu J (2012) Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochim Biophys Acta 1819:137–148
Kim KY, Kwon SY, Lee HS, Hur Y, Bang JW, Kwak SS (2003) A novel oxidative stress-inducible peroxidase promoter from sweetpotato: molecular cloning and characterization in transgenic tobacco plants and cultured cells. Plant Mol Biol 51:831–838
Kong WW, Yang ZM (2010) Identification of iron-deficiency responsive microRNA genes and cis-elements in Arabidopsis. Plant Physiol Biochem 48:153–159
Koziel MG, Beland GL, Bowman C, Carozzi NB, Crenshaw R, Crossland L, Dawson J, Desai N, Hill M, Kadwell S (1993) Field performance of elite transgenic maize plants expressing an insecticidal protein derived from Bacillus thuringiensis. Nat Biotechnol 11:194–200
Lee JT, Prasad V, Yang PT, Wu JF, David Ho TH, Chang YY, Chan MT (2003) Expression of Arabidopsis CBF1 regulated by an ABA/stress inducible promoter in transgenic tomato confers stress tolerance without affecting yield. Plant Cell Environ 26:1181–1190
Li WX, Oono Y, Zhu J, He XJ, Wu JM, Iida K, Lu XY, Cui X, Jin H, Zhu JK (2008) The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. Plant Cell 20:2238–2251
Li Y, Zhang QQ, Zhang J, Wu L, Qi Y, Zhou JM (2010) Identification of microRNAs involved in pathogen-associated molecular pattern-triggered plant innate immunity. Plant Physiol 152:2222–2231
Li X, Wang X, Zhang S, Liu D, Duan Y, Dong W (2012) Identification of soybean microRNAs involved in soybean cyst nematode infection by deep sequencing. PLoS ONE 7:e39650. doi:10.1371/journal.pone.0039650
Liang G, Yu D (2010) Reciprocal regulation among miR395, APS and SULTR2; 1 in Arabidopsis thaliana. Plant Signal Behav 5:1257–1259
Liang G, Yang F, Yu D (2010) MicroRNA395 mediates regulation of sulfate accumulation and allocation in Arabidopsis thaliana. Plant J 62:1046–1057
Liu Q, Chen YQ (2010) A new mechanism in plant engineering: the potential roles of microRNAs in molecular breeding for crop improvement. Biotechnol Adv 28:301–307
Liu HH, Tian X, Li YJ, Wu CA, Zheng CC (2008) Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA 14:836–843
Lu S (2011) miRNAs and mechanical stress. In: Wojtaszek P (ed) Mechanical induction of plant cells and plants, Springer, Heidelberg, pp 329–344
Lu S, Sun YH, Shi R, Clark C, Li L, Chiang VL (2005) Novel and mechanical stress–responsive microRNAs in Populus trichocarpa that are absent from Arabidopsis. Plant Cell 17:2186–2203
Lu S, Sun YH, Amerson H, Chiang VL (2007) MicroRNAs in loblolly pine (Pinus taeda L.) and their association with fusiform rust gall development. Plant J 51:1077–1098
Lu S, Sun YH, Chiang VL (2008) Stress-responsive microRNAs in Populus. Plant J 55:131–151
Lv DK, Bai X, Li Y, Ding XD, Ge Y, Cai H, Ji W, Wu N, Zhu YM (2010) Profiling of cold-stress-responsive miRNAs in rice by microarrays. Gene 459:39–47
Mendoza-Soto AB, Sanchez F, Hernández G (2012) MicroRNAs as regulators in plant metal toxicity response. Front Plant Sci. doi:10.3389/fpls.2012.00105
Meng Y, Shao C, Wang H, Chen M (2011) The regulatory activities of plant MicroRNAs: a more dynamic perspective. Plant Physiol 157:1583–1595
Miura K, Ikeda M, Matsubara A, Song XJ, Ito M, Asano K, Matsuoka M, Kitano H, Ashikari M (2010) OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat Genet 42:545–549
Mlotshwa S, Yang Z, Kim Y, Chen X (2006) Floral patterning defects induced by Arabidopsis APETALA2 and microRNA172 expression in Nicotiana benthamiana. Plant Mol Biol 61:781–793
Molesini B, Pii Y, Pandolfini T (2012) Fruit improvement using intragenesis and artificial microRNA. Trends Biotechnol 30:80–88
Molnar A, Bassett A, Thuenemann E, Schwach F, Karkare S, Ossowski S, Weigel D, Baulcombe D (2009) Highly specific gene silencing by artificial microRNAs in the unicellular alga Chlamydomonas reinhardtii. Plant J 58:165–174
Molnar A, Melnyk C, Baulcombe DC (2011) Silencing signals in plants: a long journey for small RNAs. Genome Biol 12:215
Moxon S, Jing R, Szittya G, Schwach F, Pilcher RLR, Moulton V, Dalmay T (2008) Deep sequencing of tomato short RNAs identifies microRNAs targeting genes involved in fruit ripening. Genome Res 18:1602–1609
Nag A, Jack T (2010) Chapter twelve—sculpting the flower; the role of microRNAs in flower development. In: Marja CPT (ed) Current topics in developmental biology, vol 91. Academic Press, New York, pp 349–378. doi:10.1016/S0070-2153(10)91012-0
Navarro L, Dunoyer P, Jay F, Arnold B, Dharmasiri N, Estelle M, Voinnet O, Jones JDG (2006) A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312:436
Navarro L, Jay F, Nomura K, He SY, Voinnet O (2008) Suppression of the microRNA pathway by bacterial effector proteins. Science 321:964–967
Ni Z, Hu Z, Jiang Q, Zhang H (2013) GmNFYA3, a target gene of miR169, is a positive regulator of plant tolerance to drought stress. Plant Mol Biol 82:113–129
Niu QW, Lin SS, Reyes JL, Chen KC, Wu HW, Yeh SD, Chua NH (2006) Expression of artificial microRNAs in transgenic Arabidopsis thaliana confers virus resistance. Nat Biotechnol 24:1420–1428
Ogita S, Uefuji H, Yamaguchi Y, Koizumi N, Sano H (2003) RNA interference: producing decaffeinated coffee plants. Nature 423:823
Ossowski S, Schwab R, Weigel D (2008) Gene silencing in plants using artificial microRNAs and other small RNAs. Plant J 53:674–690
Palatnik JF, Allen E, Wu X, Schommer C, Schwab R, Carrington JC, Weigel D (2003) Control of leaf morphogenesis by microRNAs. Nature 425:257–263
Pant BD, Musialak-Lange M, Nuc P, May P, Buhtz A, Kehr J, Walther D, Scheible WR (2009) Identification of nutrient-responsive Arabidopsis and rapeseed microRNAs by comprehensive real-time polymerase chain reaction profiling and small RNA sequencing. Plant Physiol 150:1541–1555
Perl A, Perl-Treves R, Galili S, Aviv D, Shalgi E, Malkin S, Galun E (1993) Enhanced oxidative-stress defense in transgenic potato expressing tomato Cu, Zn superoxide dismutases. Theor Appl Genet 85:568–576
Pilcher RLR, Moxon S, Pakseresht N, Moulton V, Manning K, Seymour G, Dalmay T (2007) Identification of novel small RNAs in tomato (Solanum lycopersicum). Planta 226:709–717
Pino MT, Skinner JS, Park EJ, Jeknić Z, Hayes PM, Thomashow MF, Chen THH (2007) Use of a stress inducible promoter to drive ectopic AtCBF expression improves potato freezing tolerance while minimizing negative effects on tuber yield. Plant Biotechnol J 5:591–604
Pitcher LH, Brennan E, Hurley A, Dunsmuir P, Tepperman JM, Zilinskas BA (1991) Overproduction of petunia chloroplastic copper/zinc superoxide dismutase does not confer ozone tolerance in transgenic tobacco. Plant Physiol 97:452–455
Privalle LS, Chen J, Clapper G, Hunst P, Spiegelhalter F, Zhong C (2012) Development of an agricultural biotechnology crop product: testing from discovery to commercialization. J Agric Food Chem 60:10179–10187
Puijalon S, Bouma TJ, Douady CJ, van Groenendael J, Anten NP, Martel E, Bornette G (2011) Plant resistance to mechanical stress: evidence of an avoidance-tolerance trade-off. New Phytol 191:1141–1149
Rai M, He C, Wu R (2009) Comparative functional analysis of three abiotic stress-inducible promoters in transgenic rice. Transgenic Res 18:787–799
Ramesh SV (2013) Non-coding RNAs in crop genetic modification: considerations and predictable environmental risk assessments (ERA). Mol Biotechnol 1–14. doi:10.1007/s12033-013-9648-6
Raymond Park J, McFarlane I, Hartley Phipps R, Ceddia G (2011) The role of transgenic crops in sustainable development. Plant Biotechnol J 9:2–21. doi:10.1111/j.1467-7652.2010.00565.x
Regina A, Bird A, Topping D, Bowden S, Freeman J, Barsby T, Kosar-Hashemi B, Li Z, Rahman S, Morell M (2006) High-amylose wheat generated by RNA interference improves indices of large-bowel health in rats. Proc Natl Acad Sci USA 103:3546–3551
Rissler J, Mellon MG (1996) The ecological risks of engineered crops. The MIT Press, Cambridge
Romanel E, Silva T, Corrêa R, Farinelli L, Hawkins J, Schrago CG, Vaslin MS (2012) Global alteration of microRNAs and transposon-derived small RNAs in cotton (Gossypium hirsutum) during Cotton leafroll dwarf polerovirus (CLRDV) infection. Plant Mol Biol 80:443–460
Royal Society of London (2009) Reaping the benefits: science and the sustainable intensification of global agriculture. Royal Society, London. http://royalsociety.org/uploadedFiles/Royal_Society_Content/policy/publications/2009/4294967719.pdf. Accessed 5 Feb 2013
Ru P, Xu L, Ma H, Huang H (2006) Plant fertility defects induced by the enhanced expression of microRNA167. Cell Res 16:457–465
Ruiz-Ferrer V, Voinnet O (2009) Roles of plant small RNAs in biotic stress responses. Annu Rev Plant Biol 60:485–510
Sablok G, Pérez-Quintero ÁL, Hassan M, Tatarinova TV, López C (2011) Artificial microRNAs (amiRNAs) engineering—on how microRNA-based silencing methods have affected current plant silencing research. Biochem Biophys Res Commun 406:315–319
Schramke V, Allshire R (2004) Those interfering little RNAs! Silencing and eliminating chromatin. Curr Opin Genet Dev 14:174–180
Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, Weigel D (2005) Specific effects of microRNAs on the plant transcriptome. Dev Cell 8:517–527
Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D (2006) Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18:1121–1133
Segal G, Song R, Messing J (2003) A new opaque variant of maize by a single dominant RNA-interference-inducing transgene. Genetics 165:387–397
Springer N (2010) Shaping a better rice plant. Nat Genet 42:475
Su J, Wu R (2004) Stress-inducible synthesis of proline in transgenic rice confers faster growth under stress conditions than that with constitutive synthesis. Plant Sci (Amsterdam, Neth) 166:941–948
Sun G (2012) MicroRNAs and their diverse functions in plants. Plant Mol Biol 80:17–36. doi:10.1007/s11103-011-9817-6
Sunkar R (2010) MicroRNAs with macro-effects on plant stress responses. Semin Cell Dev Biol 21:805–811
Sunkar R, Zhu JK (2004) Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 16:2001–2019
Sunkar R, Girke T, Jain PK, Zhu JK (2005) Cloning and characterization of microRNAs from rice. Plant Cell 17:1397–1411
Sunkar R, Kapoor A, Zhu JK (2006) Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell 18:2051–2065
Sunkar R, Li YF, Jagadeeswaran G (2012) Functions of microRNAs in plant stress responses. Trends Plant Sci 17:196–203
Tang G, Galili G, Zhuang X (2007) RNAi and microRNA: breakthrough technologies for the improvement of plant nutritional value and metabolic engineering. Metabolomics 3:357–369
Tang S, Wang Y, Li Z, Gui Y, Xiao B, Xie J, Zhu Q-H, Fan L (2012) Identification of wounding and topping responsive small RNAs in tobacco (Nicotiana tabacum). BMC Plant Biol 12:28
Tepperman JM, Dunsmuir P (1990) Transformed plants with elevated levels of chloroplastic SOD are not more resistant to superoxide toxicity. Plant Mol Biol 14:501–511
Tester M, Langridge P (2010) Breeding technologies to increase crop production in a changing world. Science 327:818–822. doi:10.1126/science.1183700
Thiebaut F, Rojas CA, Almeida KL, Grativol C, Domiciano GC, Lamb CRC, De Almeida Engler J, Hemerly AS, Ferreira PCG (2011) Regulation of miR319 during cold stress in sugarcane. Plant Cell Environ 35:502–512. doi:10.1111/j.1365-3040.2011.02430.x
Todesco M, Rubio-Somoza I, Paz-Ares J, Weigel D (2010) A collection of target mimics for comprehensive analysis of microRNA function in Arabidopsis thaliana. PLoS Genet 6:e1001031. doi:10.1371/journal.pgen.1001031
Tscharntke T, Clough Y, Wanger TC, Jackson L, Motzke I, Perfecto I, Vandermeer J, Whitbread A (2012) Global food security, biodiversity conservation and the future of agricultural intensification. Biol Conserv 151:53–59
Vanderschuren H, Alder A, Zhang P, Gruissem W (2009) Dose-dependent RNAi-mediated geminivirus resistance in the tropical root crop cassava. Plant Mol Biol 70:265–272
Voinnet O (2009) Origin, biogenesis, and activity of plant microRNAs. Cell 136:669–687
Wang Y, Itaya A, Zhong X, Wu Y, Zhang J, van der Knaap E, Olmstead R, Qi Y, Ding B (2011) Function and evolution of a microRNA that regulates a Ca2+-ATPase and triggers the formation of phased small interfering RNAs in tomato reproductive growth. Plant Cell 23:3185–3203
Wang S, Wu K, Yuan Q, Liu X, Liu Z, Lin X, Zeng R, Zhu H, Dong G, Qian Q (2012) Control of grain size, shape and quality by OsSPL16 in rice. Nat Genet 44:950–954
Warthmann N, Chen H, Ossowski S, Weigel D, Hervé P (2008) Highly specific gene silencing by artificial miRNAs in rice. PLoS ONE 3:e1829
World Bank (2008) World development report 2008: agriculture for development. World Bank, Washington, DC. http://siteresources.worldbank.org/INTWDR2008/Resources/WDR_00_book.pdf. Accessed 5 Feb 2013
Wu M-F, Tian Q, Reed JW (2006) Arabidopsis microRNA167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction. Development 133:4211–4218
Xie K, Wu C, Xiong L (2006) Genomic organization, differential expression, and interaction of SQUAMOSA promoter-binding-like transcription factors and microRNA156 in rice. Plant Physiol 142:280–293
Xie K, Shen J, Hou X, Yao J, Li X, Xiao J, Xiong L (2012) Gradual increase of miR156 regulates temporal expression changes of numerous genes during leaf development in rice. Plant Physiol 158:1382–1394
Xin M, Wang Y, Yao Y, Xie C, Peng H, Ni Z, Sun Q (2010) Diverse set of microRNAs are responsive to powdery mildew infection and heat stress in wheat (Triticum aestivum L.). BMC Plant Biol 10:123
Yamaguchi A, Abe M (2012) Regulation of reproductive development by non-coding RNA in Arabidopsis: to flower or not to flower. J Plant Res 125:693–704
Yang C, Li D, Mao D, Liu X, Li C, Li X, Zhao X, Chen C, Zhu L (2013) Overexpression of microRNA319 impacts leaf morphogenesis and leads to enhanced cold tolerance in rice (Oryza sativa L.). Plant Cell Environ. doi:10.1111/pce.12130
Zhang L, Hou D, Chen X, Li D, Zhu L, Zhang Y, Li J, Bian Z, Liang X, Cai X (2011a) Exogenous plant MIR168a specifically targets mammalian LDLRAP1: evidence of cross-kingdom regulation by microRNA. Cell Res 22:107–126
Zhang W, Gao S, Zhou X, Chellappan P, Chen Z, Zhou X, Zhang X, Fromuth N, Coutino G, Coffey M, Jin H (2011b) Bacteria-responsive microRNAs regulate plant innate immunity by modulating plant hormone networks. Plant Mol Biol 75:93–105
Zhang X, Zhao H, Gao S, Wang W-C, Katiyar-Agarwal S, Huang H-D, Raikhel N, Jin H (2011c) Arabidopsis argonaute 2 regulates innate immunity via miRNA393*-mediated silencing of a golgi-localized SNARE gene, MEMB12. Mol Cell 42:356–366
Zhang X, Zou Z, Gong P, Zhang J, Ziaf K, Li H, Xiao F, Ye Z (2011d) Over-expression of microRNA169 confers enhanced drought tolerance to tomato. Biotechnol Lett 33:403–409
Zhang X, Zou Z, Zhang J, Zhang Y, Han Q, Hu T, Xu X, Liu H, Li H, Ye Z (2011e) Over-expression of sly-miR156a in tomato results in multiple vegetative and reproductive trait alterations and partial phenocopy of the sft mutant. FEBS Lett 585:435–439
Zhao M, Ding H, Zhu JK, Zhang F, Li WX (2011) Involvement of miR169 in the nitrogen-starvation responses in Arabidopsis. New Phytol 190:906–915
Zhou M (2012) Genetic engineering of turfgrass for enhanced performance under environmental stress. Clemson University, Dessertation
Zhou ZS, Huang SQ, Yang ZM (2008) Bioinformatic identification and expression analysis of new microRNAs from Medicago truncatula. Biochem Biophys Res Commun 374:538–542
Zhou ZS, Zeng HQ, Liu ZP, Yang ZM (2011) Genome-wide identification of Medicago truncatula microRNAs and their targets reveals their differential regulation by heavy metal. Plant Cell Environ 35:86–99
Zhou ZS, Song JB, Yang ZM (2012) Genome-wide identification of Brassica napus microRNAs and their targets in response to cadmium. J Exp Bot 63:4597–4613
Zhou M, Li D, Li Z, Hu Q, Yang C, Zhu L, Luo H (2013) Constitutive expression of a miR319 gene alters plant development and enhances salt and drought tolerance in transgenic creeping bentgrass. Plant Physiol 161:1375–1391. doi:10.1104/pp.112.208702
Zhu X, Galili G (2003) Increased lysine synthesis coupled with a knockout of its catabolism synergistically boosts lysine content and also transregulates the metabolism of other amino acids in Arabidopsis seeds. Plant Cell 15:845–853
Zhu X, Galili G (2004) Lysine metabolism is concurrently regulated by synthesis and catabolism in both reproductive and vegetative tissues. Plant Physiol 135:129–136
Zhu QH, Helliwell CA (2011) Regulation of flowering time and floral patterning by miR172. J Exp Bot 62:487–495
Zhu QH, Upadhyaya NM, Gubler F, Helliwell CA (2009) Over-expression of miR172 causes loss of spikelet determinacy and floral organ abnormalities in rice (Oryza sativa). BMC Plant Biol 9:149. doi:10.1186/1471-2229-9-149
Acknowledgments
We thank Dr. Emerson Shipe for critically reading the manuscript. This work was supported by Biotechnology Risk Assessment Grant Program competitive grant no. 2007-33522-18489 and no. 2010-33522-21656 from the USDA National Institute of Food and Agriculture as well as the USDA grant CSREES SC-1700315 and SC-1700450. Technical Contribution No. 6123 of the Clemson University Experiment Station.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhou, M., Luo, H. MicroRNA-mediated gene regulation: potential applications for plant genetic engineering. Plant Mol Biol 83, 59–75 (2013). https://doi.org/10.1007/s11103-013-0089-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-013-0089-1