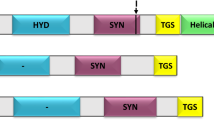
Abstract
Eukaryotic assimilatory nitrate reductase (NR) is a multi-domain protein that catalyzes the rate-limiting step in nitrate assimilation. This protein is highly conserved and has been extensively characterized in plants and algae. Here, we report hybrid NRs (NR2-2/2HbN) identified in two microalgal species, Heterosigma akashiwo and Chattonella subsalsa, with a 2/2 hemoglobin (2/2Hb) inserted into the hinge 2 region of a prototypical NR. 2/2Hbs are a class of single-domain heme proteins found in bacteria, ciliates, algae and plants. Sequence analysis indicates that the C-terminal FAD/NADH reductase domain of NR2-2/2HbN retains identity with eukaryotic NR, suggesting that the 2/2Hb domain was inserted interior to the existing NR domain architecture. Phylogenetic analysis supports the placement of the 2/2Hb domain of NR2-2/2HbN within group I (N-type) 2/2Hbs with high similarity to mycobacterial 2/2HbNs, known to convert nitric oxide to nitrate. Experimental data confirms that H. akashiwo is capable of metabolizing nitric oxide and shows that HaNR2-2/2HbN expression increases in response to nitric oxide addition. Here, we propose a mechanism for the dual function of NR2-2/2HbN in which nitrate reduction and nitric oxide dioxygenase reactions are cooperative, such that conversion of nitric oxide to nitrate is followed by reduction of nitrate for assimilation as cellular nitrogen.





Similar content being viewed by others
References
Allen AE, Ward BB, Song B (2005) Characterization of diatom (Bacillariophyceae) nitrate reductase genes and their detection in marine phytoplankton communities. J Phycol 41:95–104
Altschul SF et al (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucl Acids Res 25:3389–3402
Armbrust EV et al (2004) The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science 306:79–86
Baldauf SL (2003) The deep roots of eukaryotes. Science 300:1703–1706
Baldauf SL (2008) An overview of the phylogeny and diversity of eukaryotes. J Syst Evol 46:263–273
Berges J (1997) Miniview: algal nitrate reductases. Eur J Phycol 32:3–8
Bjorklund AK, Ekman D, Light S, Frey-Skott J, Elofsson A (2005) Domain rearrangements in protein evolution. J Mol Biol 353:911–923
Bonamore A et al (2007) A novel chimera: the “truncated hemoglobin-antibiotic monooxygenase” from Streptomyces avermitilis. Gene 398:52–61
Bowler C et al (2008) The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 456:239–244
Campbell WH (1999) Nitrate reductase structure, function and regulation: bridging the gap between biochemistry and physiology. Annu Rev Plant Physiol Plant Mol Biol 50:277–303
Campbell WH (2001) Structure and function of eukaryotic NAD(P)H:nitrate reductase. Cell Mol Life Sci 58:194–204
Chenna R (2003) Multiple sequence alignment with the clustal series of programs. Nucl Acids Res 31:3497–3500
Coyne KJ (2010) Nitrate reductase (NR1) sequence and expression in the harmful alga Heterosigma akashiwo (Raphidophyceae). J Phycol 46:135–142
Coyne KJ, Cary SC (2005) Molecular approaches to the investigation of viable dinoflagellate cysts in natural sediments from estuarine environments. J Eukaryot Microbiol 52:90–94
Coyne K, Hutchins D, Hare C, Cary S (2001) Assessing temporal and spatial variability in Pfiesteria piscicida distributions using molecular probing techniques. Aquat Microb Ecol 24:275–285
Coyne KJ, Burkholder JM, Feldman RA, Hutchins DA, Cary SC (2004) Modified serial analysis of gene expression method for construction of gene expression profiles of microbial eukaryotic species. Appl Environ Microbiol 70:5298–5304
Crawford NM (2006) Mechanisms for nitric oxide synthesis in plants. J Exp Bot 57(3):471–478
Desikan R, Griffiths R, Hancock J, Neill S (2002) A new role for an old enzyme: nitrate reductase mediated nitric oxide generation is required for abscisic acid-induced stomatal closure in Arabidopsis thaliana. PNAS 99(25)16314–16318
Doerks T, Copley RR, Shultz J, Ponting CP, Bork P (2002) Systematic identification of novel protein domain families associated with nuclear functions. Genome Res 12:47–56
Du S, Zhang Y, Lin X, Wang Y, Tang C (2008) Regulation of nitrate reductase by nitric oxide in Chinese cabbage pakchoi (Brassica chinensis L.) Plant Cell Environ 31:195–204
Eckardt NA (2005) Moco Mojo: crystal structure reveals essential features of eukaryotic assimilatory nitrate reduction. Plant Cell 17:1029–1031
Farlow A, Meduri E, Schlötterer C (2010) DNA double-strand break repair and the evolution of intron density. Trends Genet 27:1–6
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Gardner PR (2005) Nitric oxide dioxygenase function and mechanism of flavohemoglobin, hemoglobin, myoglobin and their associated reductases. J Inorg Biochem 99:247–266
Gardner PR et al (2000) Nitric-oxide dioxygenase activity and function of flavohemoglobins. Sensitivity to nitric oxide and carbon monoxide inhibition. J Biol Chem 275:31581–31587
Guillard RRL (1975) Culture of phytoplankton for feeding marine invertebrates. In: Smith WL, Chanley MH (eds) Culture of marine invertebrate animals. Plenum Press, New York, pp 26–60
Hacker J, Kaper JB (2000) Pathogenicity islands and the evolution of microbes. Annu Rev Microbiol 54:641–679
Hammer Ø, Harper DAT, Ryan PD (2001) PAST: paleontological statistics software package for education and data analysis. Palaeontologia Electronica 4:1–9
Handy SM et al (2005) Evaluating vertical migration behavior of harmful raphidophytes in the Delaware Inland Bays utilizing quantitative real-time PCR. Aquat Microb Ecol 40:121–132
Handy SM et al (2008) Using quantitative real-time PCR to study competition and community dynamis among Delaware Inland Bays harmful algae in field and laboratory studies. Harmful Algae 7:599–613
Karplus PA, Bruns CM (1994) Structure-function relations for ferredoxin reductase. J Bioenerg Biomembr 26:89–99
Kim D (2006) Nitric oxide synthase-like enzyme mediated nitric oxide generation by harmful red tide phytoplankton, Chattonella marina. J Plankton Res 28:613–620
Kim D et al (2008) Detection of nitric oxide (NO) in marine phytoplankters. J Biosci Bioeng 105:414–417
Lama A, Pawaria S, Dikshit KL (2006) Oxygen binding and NO scavenging properties of truncated hemoglobin, HbN, of Mycobacterium smegmatis. FEBS Lett 580:4031–4041
Lama A et al (2009) Role of pre-A motif in nitric oxide scavenging by truncated hemoglobin, HbN, of Mycobacterium tuberculosis. J Biol Chem 284:14457–14468
Landsberg JH (2002) The effects of harmful algal blooms on aquatic organisms. Rev Fish Sci 10:113–390
Larkin MA et al (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948
Lecomte JTJ, Vuletich DA, Lesk AM (2005) Structural divergence and distant relationships in proteins: evolution of the globins. Curr Opin Struct Biol 15:290–301
Milani M, Pesce A, Ouellet H, Guertin M, Bolognesi M (2003) Truncated hemoglobins and nitric oxide action. IUBMB Life 55:623–627
Millett F, Durham B (2004) Kinetics of electron transfer within cytochrome bc1 and between cytochrome bc1 and cytochrome c. Photosynth Res 82:1–16
Moore AD, Björklund AK, Ekman D, Bornberg-Bauer E, Elofsson A (2008) Arrangements in the modular evolution of proteins. Trends Biochem Sci 33:444–451
Mukai M, Mills CE, Poole RK, Yeh SR (2001) Flavohemoglobin, a globin with a peroxidase-like catalytic site. J Biol Chem 276:7272–7277
Nardini M, Pesce A, Milani M, Bolognesi M (2007) Protein fold and structure in the truncated (2/2) globin family. Gene 398:2–11
Oda T, Nakamura A, Okamoto T, Ishimatsu A, Muramatsu T (1998) Lectin-induced enhancement of superoxide anion production by red tide phytoplankton. Mar Biol 131:383–390
Ouellet H et al (2002) Truncated hemoglobin HbN protects Mycobacterium bovis from nitric oxide. Proc Natl Acad Sci USA 99:5902–5907
Padmakumar KB, Thomas LC, Salini TC, John E, Menon NR, Sanjeevan VN (2011) Monospecific bloom of noxious raphidophyte Chattonella marina in the coastal water of South West coast of India. Int J Biosci 1:57–59
Poole RK, Hughes MN (2000) New functions for the ancient globin family: bacterial responses to nitric oxide and nitrosative stress. Mol Microbiol 36:775–783
Rzhetsky A, Nei M (1992) Statistical properties of the ordinary least-squares, generalized least-squares, and minimum-evolution methods of phylogenetic inference. J Mol Evol 35:367–375
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sakihama Y, Nakamura S, Yamasaki H (2002) Nitric oxide production mediated by nitrate reductase in the green alga Chlamydomonas reinhardtii: an alternative NO production pathway in photosynthetic organisms. Plant Cell Physiol 43:290–297
Selvam RA, Sasidharan R (2004) DomIns: a web resource for domain insertions in known protein structures. Nucl Acids Res 32:D193–D195
Smagghe BJ, Trent JT, Hargrove MS (2008) NO dioxygenase activity in hemoglobins is ubiquitous in vitro, but limited by reduction in vivo. PloS One 3:e2039
Stolz JF, Basu P (2002) Evolution of nitrate reductase: molecular and structural variations on a common function. Chembiochem 3:198–206
Szabo C, Ischiropoulos H, Radi R (2007) Peroynitrite: biochemistry, pathophysiology and development of therapeutics. Nat Rev Drug Discov 6:662–680
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Taylor FJR, Haigh R (1993) The ecology of fish-killing blooms of the chloromonad flagellate Heterosigma in the Strait of Georgia and adjacent waters. In: Smayda TJ, Shimizu Y (eds) Toxic phytoplankton blooms in the sea. Elsevier Science Publishers, pp 705–710
Vardi A et al (2006) A stress surveillance system based on calcium and nitric oxide in marine diatoms. PLoS Biol 4(3):e60
Vardi A, Thamatrakoln K, Bidle KD, Falkowski PG (2008) Diatom genomes come of age. Genome Biol 9:245
Vinogradov SN, Moens L (2008) Diversity of globin function: enzymatic, transport, storage, and sensing. J Biol Chem 283:8773–8777
Vinogradov SN et al (2005) Three globin lineages belonging to two structural classes in genomes from the three kingdoms of life. Proc Natl Acad Sci USA 102:11385–11389
Vinogradov SN et al (2007) A model of globin evolution. Gene 398:132–142
Vuletich DA, Lecomte JTJ (2006) A phylogenetic and structural analysis of truncated hemoglobins. J Mol Evol 62:196–210
Weiner J 3rd, Beaussart F, Bornberg-Bauer E (2006) Domain deletions and substitutions in the modular protein evolution. FEBS J 273:2037–2047
Wittenberg J, Bolognesi M, Wittenberg B, Guertin M (2002) Truncated hemoglobins: a new family of hemoglobins widely distributed in bacteria, unicellular eukaryotes, and plants. J Biol Chem 277:871–874
Workun GJ, Moquin K, Rothery RA, Weiner JH (2008) Evolutionary persistence of the molybdopyranopterin-containing sulfite oxidase protein fold. Microbiol Mol Biol Rev 72:228–248
Xing L, Zhang Z-B, Liu C-Y, Wu Z–Z, Lin C (2005) Amperometric detection of nitric oxide with microsensor in the medium of seawater and its applications. Sensors 5:537–545
Yamasaki H, Sakihama Y (2000) Simultaneous production of nitric oxide and peroxynitrite by plant nitrate reductase: in vitro evidence for the NR-dependent formation of active nitrogen species. FEBS Lett 468:89–92
Yeh SR, Couture M, Ouellet Y, Guertin M, Rousseau DL (2000) A cooperative oxygen binding hemoglobin from Mycobacterium tuberculosis. Stabilization of heme ligands by a distal tyrosine residue. J Biol Chem 275:1679–1684
Zhang Z, Liu C, Wu Z, Xing L, Li P (2006) Detection of nitric oxide in culture media and studies of nitric oxide formation by marine microalgae. Med Sci Monit 12:75–86
Zhou J, Kleinhofs A (1996) Molecular evolution of nitrate reductase genes. J Mol Evol 42:432–442
Acknowledgments
This work was supported by the National Science Foundation (IOS-0745102) and the Environmental Protection Agency (STAR-ECOHAB-R83-3221). We would like to thank Lauren Salvitti, Mark Warner, and Tom Hanson (University of Delaware), and Don Stewart for technical and editorial support.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Stewart, J.J., Coyne, K.J. Analysis of raphidophyte assimilatory nitrate reductase reveals unique domain architecture incorporating a 2/2 hemoglobin. Plant Mol Biol 77, 565–575 (2011). https://doi.org/10.1007/s11103-011-9831-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-011-9831-8