Abstract
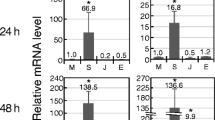
Research using the well-studied model legume Medicago truncatula has largely focused on rhizobium symbiosis, while little information is currently available for this species on pathogen-induced transcriptome changes. We have performed a transcriptome analysis of this species with the objective of studying the basal (BR, no visible symptoms) and hypersensitive response (HR, plant cell death) in its leaves at 6 and at 24 h after infection by HR-negative (hrcC mutant) and HR-inducing Pseudomonas syringae pv. syringae strains, respectively. Although there were no visible symptoms at the BR, the alterations in gene expression were comparable to those found with the HR. Both responses resulted in the transcriptional alteration of hundreds of plant genes; however, the responses in the HR were usually more intense. The reactions to HR-inducing and HR-negative bacterial strains were significantly overlapping. Parallel up- or down-regulation of genes with the same function occurred frequently. However, some plant processes were regulated in one direction; for example, most of the protein synthesis-related genes were activated and all of the photosynthetic/chloroplast genes were suppressed during BR. The possible roles of several functional classes (e.g., cell rescue, signaling, defense, cell death, etc.) of transcriptionally altered genes are discussed. The results of the comparison with available mycorrhizal and nodule expression data show that there is a significant overlap between nodulation and the leaf defense response and that during the early stage of the nodulation in roots, Sinorhizobium meliloti induces a fluctuation in the transcription of BR- and HR-responsive genes.



Similar content being viewed by others
References
Alfano JR, Collmer A (2004) Type III secretion system effector proteins: double agents in bacterial disease and plant defense. Annu Rev Phytopathol 42:385–414. doi:10.1146/annurev.phyto.42.040103.110731
Bantignies B, Seguin J, Muzac I, Dedaldechamp F, Gulick P, Ibrahim R (2000) Direct evidence for ribonucleolytic activity of a PR-10-like protein from white lupin roots. Plant Mol Biol 42:871–881. doi:10.1023/A:1006475303115
Bardwell L (2005) A walk-through of the yeast mating pheromone response pathway. Peptides 26:339–350. doi:10.1016/j.peptides.2004.10.002
Belenghi B, Acconcia F, Trovato M, Perazzolli M, Bocedi A, Polticelli F, Ascenzi P, Delledonne M (2003) AtCYS1, a cystatin from Arabidopsis thaliana, suppresses hypersensitive cell death. Eur J Biochem 270:2593–2604. doi:10.1046/j.1432-1033.2003.03630.x
Bestwick CS, Bennett MH, Mansfield JW (1995) Hrp mutant of Pseudomonas syringae pv. phaseolicola induces cell wall alterations but not membrane damage leading to the HR in lettuce (Lactuca sativa). Plant Physiol 108:503–516
Bozsó Z, Ott PG, Kecskés ML, Klement Z (1999) Effect of heat and cycloheximide treatment of tobacco on the ability of Pseudomonas syringae pv. syringae 61 hrp/hrmA mutants to cause HR. Physiol Mol Plant Pathol 55:215–223. doi:10.1006/pmpp.1999.0225
Bozsó Z, Ott PG, Szatmari A, Czelleng A, Varga G, Besenyei E, Sárdi E, Bányai E, Klement Z (2005) Early detection of bacterium-induced basal resistance in tobacco leaves with diaminobenzidine and dichlorofluorescein diacetate. J Phytopathol 153:596–607. doi:10.1111/j.1439-0434.2005.01026.x
Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC, Gaasterland T, Glenisson P, Holstege FC, Kim IF, Markowitz V, Matese JC, Parkinson H, Robinson A, Sarkans U, Schulze-Kremer S, Stewart J, Taylor R, Vilo J, Vingron M (2001) Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet 29:365–371. doi:10.1038/ng1201-365
Brechenmacher L, Weidmann S, van Tuinen D, Chatagnier O, Gianinazzi S, Franken P, Gianinazzi-Pearson V (2004) Expression profiling of up-regulated plant and fungal genes in early and late stages of Medicago truncatula-Glomus mosseae interactions. Mycorrhiza 14:253–262. doi:10.1007/s00572-003-0263-4
Breda C, Sallaud C, el-Turk J, Buffard D, de Kozak I, Esnault R, Kondorosi A (1996) Defense reaction in Medicago sativa: a gene encoding a class 10 PR protein is expressed in vascular bundles. Mol Plant Microbe Interact 9:713–719
Breitling R, Armengaud P, Amtmann A, Herzyk P (2004) Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett 573:83–92. doi:10.1016/j.febslet.2004.07.055
Carimi F, Zottini M, Formentin E, Terzi M, Lo Schiavo F (2003) Cytokinins: new apoptotic inducers in plants. Planta 216:413–421
Chen C, Wanduragala S, Becker DF, Dickman MB (2006) Tomato QM-like protein protects Saccharomyces cerevisiae cells against oxidative stress by regulating intracellular proline levels. Appl Environ Microbiol 72:4001–4006. doi:10.1128/AEM.02428-05
de Torres M, Sanchez P, Fernandez-Delmond I, Grant M (2003) Expression profiling of the host response to bacterial infection: the transition from basal to induced defence responses in RPM1-mediated resistance. Plant J 33:665–676. doi:10.1046/j.1365-313X.2003.01653.x
Deguchi Y, Banba M, Shimoda Y, Chechetka SA, Suzuri R, Okusako Y, Ooki Y, Toyokura K, Suzuki A, Uchiumi T, Higashi S, Abe M, Kouchi H, Izui K, Hata S (2007) Transcriptome profiling of Lotus japonicus roots during arbuscular mycorrhiza development and comparison with that of nodulation. DNA Res 14:117–133. doi:10.1093/dnares/dsm014
Dietz KJ (2003) Plant peroxiredoxins. Annu Rev Plant Biol 54:93–107. doi:10.1146/annurev.arplant.54.031902.134934
Dow M, Newman MA, von Roepenack E (2000) The induction and modulation of plant defense responses by bacterial lipopolysaccharides. Annu Rev Phytopathol 38:241–261. doi:10.1146/annurev.phyto.38.1.241
El Yahyaoui F, Küster H, Ben Amor B, Hohnjec N, Pühler A, Becker A, Gouzy J, Vernié T, Gough C, Niebel A, Godiard L, Gamas P (2004) Expression profiling in Medicago truncatula identifies more than 750 genes differentially expressed during nodulation, including many potential regulators of the symbiotic program. Plant Physiol 136:3159–3176. doi:10.1104/pp.104.043612
Eulgem T (2005) Regulation of the Arabidopsis defense transcriptome. Trends Plant Sci 10:71–78. doi:10.1016/j.tplants.2004.12.006
Fedorova M, van de Mortel J, Matsumoto PA, Cho J, Town CD, VandenBosch KA, Gantt JS, Vance CP (2002) Genome-wide identification of nodule-specific transcripts in the model legume Medicago truncatula. Plant Physiol 130:519–537. doi:10.1104/pp.006833
Felix G, Boller T (2003) Molecular sensing of bacteria in plants. The highly conserved RNA-binding motif RNP-1 of bacterial cold shock proteins is recognized as an elicitor signal in tobacco. J Biol Chem 278:6201–6208. doi:10.1074/jbc.M209880200
Garcia-Olmedo F, Molina A, Segura A, Moreno M (1995) The defensive role of nonspecific lipid-transfer proteins in plants. Trends Microbiol 3:72–74. doi:10.1016/S0966-842X(00)88879-4
Gelbman BD, Heguy A, O’Connor TP, Zabner J, Crystal RG (2007) Upregulation of pirin expression by chronic cigarette smoking is associated with bronchial epithelial cell apoptosis. Respir Res 8:10. doi:10.1186/1465-9921-8-10
Gomez-Gomez L, Boller T (2002) Flagellin perception: a paradigm for innate immunity. Trends Plant Sci 7:251–256. doi:10.1016/S1360-1385(02)02261-6
Greenberg JT, Yao N (2004) The role and regulation of programmed cell death in plant-pathogen interactions. Cell Microbiol 6:201–211. doi:10.1111/j.1462-5822.2004.00361.x
Guo A, Durner J, Klessig DF (1998) Characterization of a tobacco epoxide hydrolase gene induced during the resistance response to TMV. Plant J 15:647–656. doi:10.1046/j.1365-313x.1998.00241.x
Hammond-Kosack KE, Jones JDG (1996) Resistance gene-dependent plant defense responses. Plant Cell 8:1773–1791
He X, Anderson JC, del Pozo O, Gu YQ, Tang X, Martin GB (2004) Silencing of subfamily I of protein phosphatase 2A catalytic subunits results in activation of plant defense responses and localized cell death. Plant J 38:563–577. doi:10.1111/j.1365-313X.2004.02073.x
Herbers K, Mönke G, Badur R, Sonnewald U (1995) A simplified procedure for the subtractive cDNA cloning of photoassimilate-responding genes: isolation of cDNAs encoding a new class of pathogenesis-related proteins. Plant Mol Biol 29:1027–1038. doi:10.1007/BF00014975
Hirayama T, Ishida C, Kuromori T, Obata S, Shimoda C, Yamamoto M, Shinozaki K, Ohto C (1997) Functional cloning of a cDNA encoding Mei2-like protein from Arabidopsis thaliana using a fission yeast pheromone receptor deficient mutant. FEBS Lett 413:16–20. doi:10.1016/S0014-5793(97)00871-5
Hohnjec N, Vieweg MF, Puhler A, Becker A, Kuster H (2005) Overlaps in the transcriptional profiles of Medicago truncatula roots inoculated with two different Glomus fungi provide insights into the genetic program activated during arbuscular mycorrhiza. Plant Physiol 137:1283–1301. doi:10.1104/pp.104.056572
Hong JK, Hwang BK (2006) Promoter activation of pepper class II basic chitinase gene, CAChi2, and enhanced bacterial disease resistance and osmotic stress tolerance in the CAChi2-overexpressing Arabidopsis. Planta 223:433–448. doi:10.1007/s00425-005-0099-6
Hückelhoven R (2004) BAX Inhibitor-1, an ancient cell death suppressor in animals and plants with prokaryotic relatives. Apoptosis 9:299–307. doi:10.1023/B:APPT.0000025806.71000.1c
Jakobek JL, Lindgren PB (1993) Generalized induction of defense responses in bean is not correlated with the induction of the hypersensitive reaction. Plant Cell 5:49–56
Jones AM, Thomas V, Truman B, Lilley K, Mansfield JW, Grant M (2004) Specific changes in the Arabidopsis proteome in response to bacterial challenge: differentiating basal and R-gene mediated resistance. Phytochemistry 65:1805–1816. doi:10.1016/j.phytochem.2004.04.005
King EO, Ward MK, Raney DE (1954) Two simple media for the demonstration of pyocyanine and fluorescein. J Lab Clin Med 22:301–307
Klement Z (1963) Rapid detection of the pathogenicity of phytophatogenic pseudomonads. Nature 199:299–300. doi:10.1038/199299b0
Klement Z, Bozsó Z, Ott PG, Kecskés ML, Rudolp K (1999) Symptomless resistant response instead of the hypersensitive reaction in tobacco leaves after infiltration of heterologous pathovars of Pseudomonas syringae. J Phytopathol 147:467–475. doi:10.1111/j.1439-0434.1999.tb03852.x
Klement Z, Bozsó Z, Kecskés ML, Besenyei E, Czelleng A, Ott PG (2003) Local early induced resistance of plants as the first line of defence against bacteria. Pest Manag Sci 59:465–474. doi:10.1002/ps.694
Koistinen KM, Soininen P, Venalainen TA, Hayrinen J, Laatikainen R, Perakyla M, Tervahauta AI, Karenlampi SO (2005) Birch PR-10c interacts with several biologically important ligands. Phytochemistry 66:2524–2533. doi:10.1016/j.phytochem.2005.09.007
Kunze G, Zipfel C, Robatzek S, Niehaus K, Boller T, Felix G (2004) The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell 16:3496–3507. doi:10.1105/tpc.104.026765
Lapik YR, Kaufman LS (2003) The Arabidopsis cupin domain protein AtPirin1 interacts with the G protein alpha-subunit GPA1 and regulates seed germination and early seedling development. Plant Cell 15:1578–1590. doi:10.1105/tpc.011890
Liu JJ, Ekramoddoullah AKM (2006) The family 10 of plant pathogenesis-related proteins: Their structure, regulation, and function in response to biotic and abiotic stresses. Physiol Mol Plant Pathol 68:3–13. doi:10.1016/j.pmpp.2006.06.004
Liu JY, Blaylock LA, Endre G, Cho J, Town CD, VandenBosch KA, Harrison MJ (2003) Transcript profiling coupled with spatial expression analyses reveals genes involved in distinct developmental stages of an arbuscular mycorrhizal symbiosis. Plant Cell 15:2106–2123. doi:10.1105/tpc.014183
Liu J, Maldonado-Mendoza I, Lopez-Meyer M, Cheung F, Town CD, Harrison MJ (2007) Arbuscular mycorrhizal symbiosis is accompanied by local and systemic alterations in gene expression and an increase in disease resistance in the shoots. Plant J 50:529–544. doi:10.1111/j.1365-313X.2007.03069.x
Lohar DP, Sharopova N, Endre G, Penuela S, Samac D, Town C, Silverstein KA, VandenBosch KA (2006) Transcript analysis of early nodulation events in Medicago truncatula. Plant Physiol 140:221–234. doi:10.1104/pp.105.070326
Mach JM, Castillo AR, Hoogstraten R, Greenberg JT (2001) The Arabidopsis-accelerated cell death gene ACD2 encodes red chlorophyll catabolite reductase and suppresses the spread of disease symptoms. Proc Natl Acad Sci USA 98:771–776. doi:10.1073/pnas.021465298
Mackey D, McFall AJ (2006) MAMPs and MIMPs: proposed classifications for inducers of innate immunity. Mol Microbiol 61:1365–1371. doi:10.1111/j.1365-2958.2006.05311.x
Marković-Housley Z, Degano M, Lamba D, von Roepenack-Lahaye E, Clemens S, Susani M, Ferreira F, Scheiner O, Breiteneder H (2003) Crystal structure of a hypoallergenic isoform of the major birch pollen allergen Bet v 1 and its likely biological function as a plant steroid carrier. J Mol Biol 325:123–133. doi:10.1016/S0022-2836(02)01197-X
Mergaert P, Nikovics K, Kelemen Z, Maunoury N, Vaubert D, Kondorosi A, Kondorosi E (2003) A novel family in Medicago truncatula consisting of more than 300 nodule-specific genes coding for small, secreted polypeptides with conserved cysteine motifs. Plant Physiol 132:161–173. doi:10.1104/pp.102.018192
Murray GI, Paterson PJ, Weaver RJ, Ewen SW, Melvin WT, Burke MD (1993) The expression of cytochrome P-450, epoxide hydrolase, and glutathione S-transferase in hepatocellular carcinoma. Cancer 71:36–43. doi:10.1002/1097-0142(19930101)71:1<36::AID-CNCR2820710107>3.0.CO;2-J
Mysore KS, Crasta OR, Tuori RP, Folkerts O, Swirsky PB, Martin GB (2002) Comprehensive transcript profiling of Pto- and Prf-mediated host defense responses to infection by Pseudomonas syringae pv. tomato. Plant J 32:299–315. doi:10.1046/j.1365-313X.2002.01424.x
Nakashita H, Yasuda M, Nitta T, Asami T, Fujioka S, Arai Y, Sekimata K, Takatsuto S, Yamaguchi I, Yoshida S (2003) Brassinosteroid functions in a broad range of disease resistance in tobacco and rice. Plant J 33:887–898. doi:10.1046/j.1365-313X.2003.01675.x
Navarro L, Zipfel C, Rowland O, Keller I, Robatzek S, Boller T, Jones JD (2004) The transcriptional innate immune response to flg22. Interplay and overlap with AVR gene-dependent defense responses and bacterial pathogenesis. Plant Physiol 135:1113–1128. doi:10.1104/pp.103.036749
Newman JW, Morisseau C, Hammock BD (2005) Epoxide hydrolases: their roles and interactions with lipid metabolism. Prog Lipid Res 44:1–51. doi:10.1016/j.plipres.2004.10.001
Novacky A (1972) Suppression of the bacterially induced hypersensitive reaction by cytokinins. Physiol Plant Pathol 2:101–104. doi:10.1016/S0048-4059(72)80004-3
Oh HS, Kwon H, Sun SK, Yang CH (2002) QM, a putative tumor suppressor, regulates proto-oncogene c-yes. J Biol Chem 277:36489–36498. doi:10.1074/jbc.M201859200
Orzaez D, de Jong AJ, Woltering EJ (2001) A tomato homologue of the human protein PIRIN is induced during programmed cell death. Plant Mol Biol 46:459–468. doi:10.1023/A:1010618515051
Ott PG, Szabó L, Balázs E, Klement Z (1997) Submicroscopic evidence of bacterially induced resistance in tobacco leaves. Acta Phytopathol Entomol Hung 32:265–280
Ott PG, Varga GJ, Szatmari A, Bozsó Z, Klement E, Medzihradszky KF, Besenyei E, Czelleng A, Klement Z (2006) Novel extracellular chitinases rapidly and specifically induced by general bacterial elicitors and suppressed by virulent bacteria as a marker of early basal resistance in tobacco. Mol Plant Microbe Interact 19:161–172. doi:10.1094/MPMI-19-0161
Pang H, Bartlam M, Zeng Q, Miyatake H, Hisano T, Miki K, Wong LL, Gao GF, Rao Z (2004) Crystal structure of human pirin: an iron-binding nuclear protein and transcription cofactor. J Biol Chem 279:1491–1498. doi:10.1074/jbc.M310022200
Pastuglia M, Swarup R, Rocher A, Saindrenan P, Roby D, Dumas C, Cock JM (2002) Comparison of the expression patterns of two small gene families of S genefamily receptor kinase genes during the defence response in Brassica oleracea and Arabidopsis thaliana. Gene 282:215–225. doi:10.1016/S0378-1119(01)00821-6
Pinot F, Bosch H, Alayrac C, Mioskowski C, Vendais A, Durst F, Salaun JP (1993) [omega]-hydroxylation of oleic acid in Vicia sativa microsomes (inhibition by substrate analogs and inactivation by terminal acetylenes). Plant Physiol 102:1313–1318
Ruiz-Lozano JM, Roussel H, Gianinazzi S, Gianinazzi-Pearson V (1999) Defense genes are differentially induced by a mycorrhizal fungus and Rhizobium sp. in wild-type and symbiosis-defective pea genotypes. Mol Plant Microbe Interact 12:976–984. doi:10.1094/MPMI.1999.12.11.976
Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J (2003) TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34:374–378
Sanmartin M, Jaroszewski L, Raikhel NV, Rojo E (2005) Caspases. Regulating death since the origin of life. Plant Physiol 137:841–847. doi:10.1104/pp.104.058552
Schweighofer A, Hirt H, Meskiene I (2004) Plant PP2C phosphatases: emerging functions in stress signaling. Trends Plant Sci 9:236–243. doi:10.1016/j.tplants.2004.03.007
Sheehan D, Meade G, Foley VM, Dowd CA (2001) Structure, function and evolution of glutathione transferases: implications for classification of non-mammalian members of an ancient enzyme superfamily. Biochem J 360:1–16. doi:10.1042/0264-6021:3600001
Siciliano V, Genre A, Balestrini R, Cappellazzo G, deWit PJ, Bonfante P (2007) Transcriptome analysis of arbuscular mycorrhizal roots during development of the prepenetration apparatus. Plant Physiol 144:1455–1466. doi:10.1104/pp.107.097980
Sikorski MM, Biesiadka J, Kasperska AE, Kopciska J, Lotocka B, Golinowski W, Legocki AB (1999) Expression of genes encoding PR10 class pathogenesis-related proteins is inhibited in yellow lupine root nodules. Plant Sci 149:125–137. doi:10.1016/S0168-9452(99)00148-X
Stuhlfelder C, Mueller MJ, Warzecha H (2004) Cloning and expression of a tomato cDNA encoding a methyl jasmonate cleaving esterase. Eur J Biochem 271:2976–2983. doi:10.1111/j.1432-1033.2004.04227.x
Szatmari A, Ott PG, Varga GJ, Besenyei E, Czelleng A, Klement Z, Bozso Z (2006) Characterisation of basal resistance (BR) by expression patterns of newly isolated representative genes in tobacco. Plant Cell Rep 25:728–740. doi:10.1007/s00299-005-0110-5
Takemoto D, Yoshioka H, Doke N, Kawakita K (2003) Disease stress-inducible genes of tobacco: expression profile of elicitor-responsive genes isolated by subtractive hybridization. Physiol Plant 118:545–553. doi:10.1034/j.1399-3054.2003.00145.x
Taler D, Galperin M, Benjamin I, Cohen Y, Kenigsbuch D (2004) Plant R genes that encode photorespiratory enzymes confer resistance against disease. Plant Cell 16:172–184. doi:10.1105/tpc.016352
Tao Y, Xie Z, Chen W, Glazebrook J, Chang HS, Han B, Zhu T, Zou G, Katagiri F (2003) Quantitative nature of Arabidopsis responses during compatible and incompatible interactions with the bacterial pathogen Pseudomonas syringae. Plant Cell 15:317–330. doi:10.1105/tpc.007591
Tellström V, Usadel B, Thimm O, Stitt M, Küster H, Niehaus K (2007) The lipopolysaccharide of Sinorhizobium meliloti suppresses defense-associated gene expression in cell cultures of the host plant Medicago truncatula. Plant Physiol 143:825–837. doi:10.1104/pp.106.090985
Thilmony R, Underwood W, He SY (2006) Genome-wide transcriptional analysis of the Arabidopsis thaliana interaction with the plant pathogen Pseudomonas syringae pv. tomato DC3000 and the human pathogen Escherichia coli O157:H7. Plant J 46:34–53. doi:10.1111/j.1365-313X.2006.02725.x
Truman W, de Zabala MT, Grant M (2006) Type III effectors orchestrate a complex interplay between transcriptional networks to modify basal defence responses during pathogenesis and resistance. Plant J 46:14–33. doi:10.1111/j.1365-313X.2006.02672.x
Wendler WM, Kremmer E, Förster R, Winnacker EL (1997) Identification of pirin, a novel highly conserved nuclear protein. J Biol Chem 272:8482–8489. doi:10.1074/jbc.272.43.27091
Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) Genevestigator. Arabidopsis microarray database and analysis toolbox. Plant Physiol 136:2621–2632. doi:10.1104/pp.104.046367
Acknowledgments
This research was supported by grants of the Hungarian National Science Foundation, AT-049318 and K68386. We thank the GODMAP platform (Gif-sur-Yvette, France) for the use of their microarray facilities.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Bozsó, Z., Maunoury, N., Szatmari, A. et al. Transcriptome analysis of a bacterially induced basal and hypersensitive response of Medicago truncatula . Plant Mol Biol 70, 627–646 (2009). https://doi.org/10.1007/s11103-009-9496-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-009-9496-8