Abstract
Glutamate is the major excitatory neurotransmitter in the mammalian central nervous system. After its release from presynaptic nerve terminals, glutamate is quickly removed from the synaptic cleft by excitatory amino acid transporters (EAATs) 1–5, a subfamily of glutamate transporters. The five proteins utilize a complex transport stoichiometry that couples glutamate transport to the symport of three Na+ ions and one H+ in exchange with one K+ to accumulate glutamate against up to 106-fold concentration gradients. They are also anion-selective channels that open and close during transitions along the glutamate transport cycle. EAATs belong to a larger family of secondary-active transporters, the SLC1 family, which also includes purely Na+- or H+-coupled prokaryotic transporters and Na+-dependent neutral amino acid exchangers. In recent years, molecular cloning, heterologous expression, cellular electrophysiology, fluorescence spectroscopy, structural approaches, and molecular simulations have uncovered the molecular mechanisms of coupled transport, substrate selectivity, and anion conduction in EAAT glutamate transporters. Here we review recent findings on EAAT transport mechanisms, with special emphasis on the highly conserved hairpin 2 gate, which has emerged as the central processing unit in many of these functions.
Similar content being viewed by others
Transport Functions of EAAT Glutamate Transporters
The excitatory amino acid transporters (EAATs) are the predominant glutamate transporters in the mammalian brain [1]. They were first studied in detail by Baruch Kanner and colleagues in experiments using radiotracer flux experiments and membrane vesicles from the mammalian brain [2,3,4,5]. In his seminal work, Baruch described the key features of EAAT function: stereoselective glutamate uptake and the coupling of glutamate transport to the inward movement of at least two Na+, with obligatory K+ dependence. He also established the first kinetic transport model [5], containing distinct Na+ and K+ hemicycles; it is still in use, with only minor modifications.
Baruch headed one of the three groups that almost simultaneously identified the amino acid sequences of major glutamate transporters. In 1992, Stoffel and colleagues [6] reported the amino acid sequence of a glycoprotein (GLAST) that was co-purified during the isolation of the UDP-galactose:ceramide galactosyltransferase from rat brain. The protein sequence was similar to those of bacterial transport proteins, and heterologous expression in Xenopus oocytes resulted in radioactive glutamate uptake by injected oocytes. Two weeks later, Baruch´s group reported the isolation of a complementary DNA clone encoding the glial glutamate transporter GLT-1 [7], and Kanai and Hediger reported expression cloning of EAAC1 from rabbit small intestine [8]. Subsequently, Amara and colleagues [9] cloned the three human homologs and introduced the EAAT nomenclature by naming them EAAT1, EAAT2, and EAAT3. Homology cloning of EAAT4 cDNA from the cerebellum [10] and of EAAT5 cDNA from the retina [11] completed the mammalian EAAT family. Identification of the protein sequences was the first step toward understanding EAAT function at the molecular level and permitted heterologous expression and high-resolution electrical recording of EAAT currents, genetic modification in animal models, and the linkage of glutamate transporter dysfunction to inherited diseases.
Voltage clamp analysis of injected Xenopus oocytes or transfected mammalian cells provided insights into EAAT function at unprecedented accuracy [12,13,14,15]. Comparison of glutamate-elicited currents in oocytes expressing EAAT1, EAAT2, or EAAT3 [16] led to the discovery of an EAAT-mediated anion current that is not thermodynamically coupled to amino acid transport. EAAT anion currents largely exceeded transport currents in EAAT4 and EAAT5 [10, 11, 17, 18]. Noise analysis of glutamate-activated current in isolated photoreceptors from the tiger salamander estimated unitary currents that were well above the limits of transporter-mediated fluxes [19], thus providing compelling evidence that EAATs are not only secondary-active glutamate transporters, but can also function as anion channels [20].
In electrogenic transporters, the transport stoichiometry predicts the current reversal potential as a function of the intra- and extracellular ion concentrations. Measurements of transport reversal potentials under various ionic conditions unambiguously demonstrated that EAAT1, EAAT2, and EAAT3 transport three Na+ and one H+ along with glutamate, in exchange for one K+ [13, 21, 22], indicating that EAATs have the most complex transport stoichiometry of all secondary active transporters. A comparison with prokaryotic homologs that couple three Na+ to transport of the amino acid substrate [23] illustrates that K+ coupling was a late evolutionary addition; however, the physiological importance of this optimization mechanism remains unclear. Astrocytes are most important for glutamate uptake in the mammalian brain. The resting potential of astrocytes is the K+ equilibrium potential, at which the driving forces for K+-coupled and K+-independent transporters are identical. Notably, K+ coupling increases the driving force in cells that have membrane potentials positive to the K+ equilibrium potential.
Cellular Roles of Excitatory Amino Acid Transporters
Mammalian EAATs transport glutamate into glial, neuronal, and epithelial cells. EAAT1 and EAAT2 are mainly expressed in glial cells [24]; however, recent data demonstrated that EAAT2 is also present in presynaptic nerve terminals [25, 26]. Whereas the genetic ablation of EAAT2/GLT-1 in mice resulted in lethal spontaneous seizures [27], EAAT1/GLAST knockout led only to motor discoordination and increased susceptibility to cerebellar injury, illustrating a less important role of this isoform in glutamate homeostasis [28]. EAAT3/EAAC1 is not only expressed in neuronal cells, but also in epithelial cells, in which it serves as glutamate uptake system in the kidney and the gastrointestinal tract. Mice lacking EAAT3/EAAC1 exhibit dicarboxylic aminoaciduria owing to impaired amino acid uptake in the proximal tubule [29]. EAAT4 is expressed predominantly in the cerebellum, where it seems to regulate Purkinje cell activity and motor behavior [30]. EAAT5 has been proposed to function as a presynaptic glutamate-gated chloride channel in the retina, where it mediates a negative feedback mechanism for glutamate release [31, 32]. However, no EAAT5 knockout animal has yet been reported. Taken together, these experiments demonstrate that EAAT2 is indispensable for extracellular glutamate homeostasis in the mammalian brain, whereas the other isoforms have only modulatory functions.
The linkage of human genetic diseases to genes encoding EAAT1 (SLC1A3), EAAT2 (SLC1A2) and EAAT3 (SLC1A1) revealed additional roles of EAATs in cell and organ physiology. Mutations in SLC1A3 cause episodic ataxia type 6 (EA6), a genetic condition characterized by ataxia, epilepsy and hemiplegia. The first reported patient with a disease-associated SLC1A3 mutation was found to be heterozygous for a proline to arginine substitution at position 290 (P290R) of EAAT1 [33]. This amino acid substitution impairs the glutamate transport rate, but increases anion channel activity in heterologous expression systems [34, 35]. In a transgenic mouse model (Slc1a3P290R/+) that has neurological symptoms closely resembling the human disease, ataxia is caused by Bergmann glia apoptosis during infancy and consequent cerebellar degeneration [36]. Bergmann glia apoptosis is triggered by cell shrinking due to increased glutamate-activated Cl− efflux and is thus a direct consequence of gain of function in the P290R EAAT1 anion channel. EAAT1 anion channels appear to be a main determinant in setting the resting [Cl−] in Bergmann glia cells of wildtype (WT) mice [37]. Although enhanced anion channel function is the main factor in EA6 pathogenesis in this particular case, a later analysis did not reveal similar functional changes for seven other SLC1A3 mutations in EA6 patients [38]. In addition to episodic ataxia, migraine [39], Tourette syndrome [40], and attention deficit hyperactivity disorder and autism [41] have been associated with SLC1A3 mutations.
SLC1A2 mutations that predict amino acid exchanges G82R, L85P, or P289R in EAAT2 were reported in three extraordinarily severe cases of epilepsy [42, 43]. The functional consequences of these mutations have not been studied; thus, it is unclear how these mutations affect EAAT2 functions and cause hyperexcitability in the affected individuals. Lastly, two cases of human dicarboxylic aminoaciduria were caused by mutations in SLC1A1: a three base pair deletion (c.1184–1186delTCA) and a base pair exchange causing the R445W mutation in EAAT3 [44]. These disease-causing SLC1A1 mutations illustrate the importance of EAAT3 for amino acid reabsorption in the kidney.
Structural Basis of Glutamate Transport
The first three-dimensional structure of a SLC1 transporter was determined for GltPh, an EAAT homolog of the prokaryote Pyrococcus horikoshii [45]. GltPh is assembled as a trimer, with each protomer consisting of eight transmembrane helices (TMs) and two helical hairpin loops (HP1 and HP2). Each protomer contains two distinct domains: the trimerization domain, which mediates inter-subunit interactions, and the transport domain, which harbors the ion- and substrate binding sites (Fig. 1a). Crystal and cryo-electron microscopy structures of GltPh and of another prokaryotic homolog GltTk [45,46,47,48,49,50,51,52] captured the transporter in multiple conformations. For GltPh, these include the substrate-bound [45], the TBOA-bound [46] and the Na+-only bound [49] outward-facing conformation, an intermediate conformation [51], and the substrate-bound inward-facing conformation [50]. Crystal GltTk structures were the first to reveal the apo [47] and the fully bound [52] outward-facing conformations, which defined the substrate-binding pocket and all Na+-binding sites (including the elusive Na3 binding site). Furthermore, cryo-EM provided GltTk structures in various conformational states, including a Na+-only bound inward-facing [48] state. The substrate binds between the tips of HP1 and HP2: the different structures exhibit marked variability in the position of HP2, illustrating ion-induced conformational rearrangements of this structural element (Fig. 1b).
Structures of outward- and inward-facing conformations of GltPh. a Structure of a GltPh trimer (PDB ID: 4OYE) in top view (surface, trimerization domains; cartoon, transport domains with HP1 and HP2 highlighted in gray and black/violet, respectively). b Close-up of the binding pocket with HP2 in the closed (violet, PDB ID: 4OYE), intermediate (teal, PDB ID: 4OYF), and open (orange, PDB ID: 7AHK) conformations. c, d GltPh outward-facing (OFC; PDB ID: 2NWX) (c) and inward-facing (IFC; PDB ID: 3KBC). d conformations in the fully-bound (Na1–3) state in sideview (pink spheres, bound aspartate; green spheres, Na+ ions). The Na3-bound ion was not resolved in these structures, and its position was inferred from the Na+-only bound GltPh structure (PDB ID: 7AHK)
A comparison of GltPh structures in outward-facing (OFC) [45, 46], inward-facing (IFC) [50], and intermediate (IC) [51] conformations revealed that large-scale (~ 18 Å) rotational/translational movements of the transport domain along the trimerization domain are the basis of substrate and Na+ transmembrane ion transport (Fig. 1c, d). Earlier bioinformatic analyses predicted correctly that such elevator rigid-body transmembrane motion of a mobile domain relative to a static domain is the basis of alternating accessibility [53]. Recently, the crystal structure of thermostabilized human EAAT1 [54] and the cryo-electron microscopy structures of the alanine/serine/cysteine/threonine transporter ASCT2 in several conformations [55,56,57] illustrated structural conservation of the GltX fold during evolution and confirmed the validity of the elevator transport mechanism.
HP2 Flexibility is a Key Determinant of Na+-Substrate Coupling in Glutamate Transporters
HP2 controls accessibility of the ligand-binding pocket as an extracellular and intracellular gate [46, 57]. Since its open conformation is sterically incompatible with translocation of the transport domain [46, 47, 52, 58], the opening–closing transitions of HP2 also control the translocation process [48, 58]. The competitive inhibitor TBOA blocks transport by locking the transporter in an OFC with an open HP2 [46, 54] and reduces the occupation time of the intermediate state [59].
Glutamate uptake is coupled to the symport of three Na+, making the [Na+] gradient the most important driving force for synaptic glutamate clearance. The three Na+ bind to three different Na+-binding sites: Na1 is located below the substrate-binding pocket and thus needs to be occupied prior to substrate binding, whereas Na2 is formed by the closed HP2 and TM7 and can only be occupied upon the closure of HP2 after substrate binding [46]. The third Na+-binding site, Na3, was initially proposed in computational studies [60] and recently identified in a crystal structure of fully-bound GltTk [52] and in a GltPh structure in the Na+-only bound state [49].
In unguided atomistic molecular dynamics (MD) simulations of the outward-facing apo GltPh, HP2 was found to be intrinsically flexible, resulting in spontaneous transitions between the open and closed conformations (Fig. 2a) [49]. In these simulations, Na+ binds spontaneously after HP2 opening and subsequent hydration of the Na1 site (Fig. 2b). Occupation of the Na3 site was not observed in unguided MD simulations on timescales up to 10 µs, suggesting that Na3 association is rate-limiting for Na+-induced conformation changes (see below, Fig. 3).
Na+-dependent gate dynamics in GltPh. a, b Time courses of HP2 opening (measured as the distance between S279 and G354, located on the tips of HP1 and HP2, respectively) (a) and of the distance of the closest Na+ ion from the Na1 site (b) in a representative all-atom MD simulation of GltPh in an outward-facing, gate-closed apo state. c The kobs for rapid Na+ application to detergent-solubilized F273W GltPh in stopped-flow fluorescence experiments at 20 °C and corresponding fits of the kinetic model described in (e). d Probability densities for HP2 opening as a function of Na1- and Na3-site occupancies, as obtained by umbrella sampling simulations (EV, eigenvector). e Four-state model describing Na+ binding to the apo transporter. T and T* indicate the closed and open states of HP2, respectively. Figure partially reprinted from [49]
Induced fit substrate association with GltPh. a Tryptophan fluorescence transients upon rapid mixing of 500 µM l-aspartate to L130W GltPh pre-equilibrated with 250 mM NaCl (upper curve) or upon simultaneous mixing of 500 µM l-aspartate and 250 mM NaCl with empty L130W GltPh (lower curve). NaCl was substituted with equimolar ChoCl. Colored lines are experimental data, and fits to sums of exponential functions are in gray. b Tryptophan fluorescence transients after rapid mixing of L130W GltPh in 200 mM NaCl with various [l-aspartate]. Experimental data are in colors, and fits to the sum of two exponential functions or to a linear function (trace of buffer alone) are in gray. For each trace, the fitted value of kobs is given. c Amino acid concentration dependence of kobs, obtained as shown in (b). d Left: Time course of 3H-l-aspartate (100 nM) uptake, as driven by an inwardly directed [Na+] gradient in the absence or presence of l-cysteine sulfinic acid (l-CS) and l-aspartate (10 µM each). The first 120 s of a representative experiment are shown, with GltPh-free liposomes as control. e Initial uptake rates in the absence or presence of competitor (mean ± SEM, three experiments). Figure partially reprinted from [64]
WT GltPh completely lacks tryptophan residues, and tryptophan insertion at position 273 enabled conformational changes upon Na+ binding to be followed using tryptophan fluorescence spectroscopy [61]. W273 reports on the association of Na+ and aspartate with opposing changes in fluorescence intensity. Association with Na+ and aspartate have opposite effects on HP2 [62]. This finding, together with the differential water accessibility of this residue observed in MD simulations [49], demonstrates that W273 directly reports on HP2 opening and closing. Fast stopped-flow application of Na+ to solubilized F273W GltPh triggers an increase in fluorescence, with observed rate constant (kobs) decreasing as the [Na+] rises to ~ 50 mM and then increasing to a plateau at higher Na+ concentrations (Fig. 2c). The initial decrease in the kobs demonstrates that a Na+-independent conformational change occurs before Na+ binding, which resembles a conformational selection mechanism [63]. Subsequent acceleration of kobs with increasing [Na+] reveals an additional Na+-dependent process. Therefore, Na+ binding to apo GltPh can be described by a state diagram in which initial HP2 gate opening is followed by binding to Na1 and Na3.
The Na+ dependence of HP2 opening rates provides experimental evidence that occupation of the Na1 and Na3 sites modifies the energetics of HP2 opening. Umbrella sampling simulations demonstrated that HP2 assumes mostly an intermediate conformation in apo GltPh, and that Na+ binding to only the Na1 site partially promotes the open states, while occupation of both the Na1 and Na3 sites results in almost exclusive adoption of the open state (Fig. 2d). A comparison of apo, Na+-bound, and Na+/l-aspartate-bound GltPh crystal structures revealed different orientations of residues around the Na+-binding sites, indicating that these residues undergo a series of rearrangements during Na+ association. Thus, Na+ association starts with the spontaneous opening of HP2, followed by a series of discrete conformational changes that result in maturation of the Na1 and Na3 binding sites: subsequent binding of Na+ to Na1 and Na3 stabilizes HP2 in an open state (Fig. 2e).
HP2 Closure Ensures High Substrate Selectivity in Glutamate Transporters
The flexibility of HP2 allows for exposure of the substrate-binding pocket in the apo conformation; thus, aspartate binding might be possible without prior Na+ association. However, experimentally obtained EAAT/GltX transport stoichiometries are constant over a large concentration range [13], demonstrating the existence of mechanisms that prevent substrate association to the Na+-free transporter. In unguided MD simulations at physiological [NaCl] and [Asp−], aspartate binding also occurred in apo and Na1-bound GltPh; however, Na+ occupation of both Na1 and Na3 strongly increased aspartate densities near the aspartate-binding site. These results suggest that electrostatic attraction of aspartate by bound Na+ ions effectively stimulates substrate association [49].
HP2 closes after l-aspartate association [45, 61, 64, 65], and tryptophan fluorescence spectroscopy revealed an induced fit mechanism of aspartate binding [64]. Figure 3a shows fluorescence changes upon mixing of L130W GltPh with 500 µM l-aspartate and 250 mM Na+, as compared to application of l-aspartate to L130W GltPh pre-incubated with Na+ at the same concentrations. Whereas simultaneous application of l-aspartate and Na+ results in slow multi-exponential fluorescence increases, addition of l-aspartate alone to the transporter pre-incubated with Na+ caused fluorescence changes proceeding with one rate, that is substantially faster than the largest rate observed upon simultaneous addition of Na+ and l-aspartate to the apo transporter. The observed rate constants of this process (kobs) increase with [l-aspartate] in a saturable fashion (Fig. 3c). This behavior reflects the closure of HP2 after aspartate association, which is the key determinant of substrate selectivity in EAATs/Glts. l-cysteine sulfinic acid binds with a higher affinity than l-aspartate (Fig. 3c) but is transported less effectively (Fig. 3d and e) due to slower closure of HP2 upon l-cysteine sulfinic acid binding compared with l-aspartate binding (Fig. 3c). In contrast, d-aspartate-binding rates saturate at higher values compared with l-aspartate and l-cysteine sulfinic acid (Fig. 3c). Dependence of the HP2 closure rate on the bound substrate establishes that GltPh binds amino acids via an induced fit mechanism [64]. This mechanism permits the preferential transport of l-aspartate, which binds with a lower affinity than other substrates and is then released easily on the other membrane side. An induced fit mechanism is thus responsible for high selectivity combined with high effectivity in glutamate transporters.
Changes in HP2 Dynamics Confer Obligate K+ Coupling to EAATs
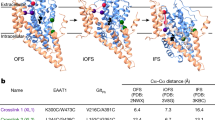
K+-dependent re-translocation is the final step of the glutamate transport cycle and confers obligatory K+ dependence on mammalian EAATs [4, 5]. Since prokaryotic transporters are K+ independent [66, 67], an EAAT-specific K+-binding site has been assumed—for many years—to have developed late in evolution. Surprisingly, in unbiased MD simulations K+ binds to multiple sites in both GltPh and EAAT1 [68, 69]. Three of these binding sites (K1, K2, and K3) are identical in GltPh and EAAT1, and only K4 is EAAT1-specific (Fig. 4a). However, only K1 and K2 are of sufficiently high affinity and selectivity to serve as binding sites for K+-bound translocation: K1 has higher affinity, but lower selectivity than K2 (Fig. 4b) [68]. In simulations, the transport domain with K+ bound to K1 carries a negative charge during translocation, whereas occupation of K2 induces the opposite charge movement [68]. In experiments, fast application of K+ resulted in the inward translocation of negative charges (Fig. 4c) [70], demonstrating that K1, and not K2, is occupied during translocation in EAATs.
Allosteric gate modulation by K+. a Isodensity meshes showing the K+ density (contoured at 3.5σ) around an EAAT1 monomer in the outward-facing conformation. b Left: EAAT1 K+ dissociation constants from unguided MD simulations, as obtained by calculating the off rates by second-order rate constants of ion binding. Right: Na+/K+ selectivities, as calculated from alchemical free-energy simulations. c Current responses to a rapid solution exchange from 140 mM CholGluc to 140 mM KGluc for HEK293T cells expressing WT EAAT1 (n = 3). Cells were internally dialyzed with a 115 mM KGluc-based solution and held at 0 mV. d Probability density distribution obtained from umbrella sampling simulations for HP2 opening in WT and L448A EAAT1; closed states are indicated by the shaded area. e Representative current recordings from cells expressing L448A or WT EAAT1 upon voltage steps to -140 mV before and after superfusion with 1 mM l-glutamate. Cells were internally dialyzed with a K+-free solution. f Net transport current amplitudes for WT and L448A EAAT1 for different intracellular solutions (***p < 0.001, Student's t‐test). Figure partially reprinted from [68]
Since K+-independent GltPh contains the same K+-binding sites as obligatory K+-coupled EAATs, other mechanisms must be responsible for the distinct transport stoichiometries of mammalian and prokaryotic transporters. In the prokaryotic transporter GltPh, HP2 can close in the apo state [47], so that the transport domain can translocate without bound K+ in the apo state. In contrast, HP2 is almost always open in apo EAAT1, and K+ binding is required for EAAT1 HP2 closure and transmembrane translocation. These finding demonstrate that K+-independent and obligatory K+-coupled transporters differ in their HP2 gate dynamics and predict that mutations in HP2, that render the closed state without bound K+ energetically more favorable, can convert K+-coupled transporters into K+-independent transporters. Figure 4d illustrates HP2 dynamics of one of such mutations, L448A EAAT1, in umbrella sampling simulations. In this mutant, HP2 closure also occurs in the absence of bound K+. In experiments, L448A EAAT1 transports glutamate also in the absence of K+, demonstrating that changes in HP2 dynamics are sufficient for conferring K+ independence to EAAT1 transporters [68] (Fig. 4e).
As the Na+- and K+-binding sites partially overlap, K+ has to be released before Na+ can bind. K+ cannot dissociate directly from K1 to the bulk solution when HP2 is closed. However, the intrinsic flexibility of HP2 allows for opening with low probability in presence of K+ bound to K1, thus permitting K+ to dissociate (Fig. 4d). Moreover, K+ can move from K1 to K2 and dissociate to the external medium without prior HP2 opening [68]. HP2 opens after K+ dissociation, permitting Na+ association to Na1 and Na3.
Differences in K+ coupling in the EAATs/GltX are based on distinct allosteric K+–HP2 interactions [68, 71], illustrating how allosteric coupling permits modification of transport stoichiometries without the formation of novel binding sites [72, 73]. It may also account for the related ASCTs merely functioning as electroneutral exchangers of neutral amino acids. Small sequence differences in these isoforms may further increase the open probability of HP2 in the apo state, such that HP2 closure would only be possible after the binding of Na+ and amino acids, thereby converting coupled vectorial transporters into obligatory exchangers [74].
EAAT Anion Channels Open via Lateral Movement of the Transport Domain
EAATs/GltX are not only glutamate transporters, but also anion channels that open in response to transitions within the glutamate transport cycle [16, 75,76,77,78]. Atomistic MD simulations permitted the identification of an anion-conducting channel conformation [79] that accounts for the experimentally observed lyotropic anion selectivity [14, 80] and unitary current amplitudes in EAATs [81, 82].
In all-atom MD simulations of GltPh, the OFC and IFC turned out to be non-conductive to ions, with no Cl− permeation even in presence of high transmembrane voltages [79] (Fig. 5a). Furthermore, intermediate conformations—either obtained by X-ray crystallography [51] or by enhanced-sampling simulations of transmembrane translocation [79]—also showed no ion permeation. However, extended unguided simulations starting from such translocation intermediates sampled reversible transitions to a channel-like conformation (ChC) defined by a lateral movement of the mobile substrate transport domain. Transitions to the ChC involved the opening of a cleft between the transport and trimerization domains that permitted frequent Cl− permeation events with partially retained hydration shell after water influx (Fig. 5b and c). Hundreds of simulated anion permeation events defined the conduction pathway with a relatively wide anion pore (minimum diameter of 5.6 Å) and large vestibules on both membrane sites in the ChC (Fig. 5d). Simulations with either NaCl and NaI solutions revealed perfect anion-over-cation selectivity and larger conductance for I– than for Cl− (Fig. 5e). Large anions as glutamate or aspartate were impermeant, and simulated anion permeation rates were consistent with experimental EAAT unitary anion current amplitudes determined by noise analysis. Several amino acids that line the anion pore are hydrophobic, with only one positively charged pore-forming residue, arginine R276 at the tip of HP1. EAATs lack a positive side chain at the position corresponding to R276, but contain an arginine at the position corresponding to M395 in TM8 of GltPh. This arginine projects its side chain into the same location in the anion conduction pathway and fulfills the same functional role.
Anion conduction in glutamate transporters. a Simulated averaged chloride densities (teal mesh) contoured at 0.2σ at the interface between the transport and trimerization domain in the intermediate conformation (IC; in-between the OFC and IFC) of GltPh, side view. b Conformational changes during anion pore formation, shown as superposition of the IC and open-channel conformation (ChC) onto the static trimerization domains (black) in top view (light gray, transport domain of the IC; teal, transport domain of the ChC) upon application of a membrane potential. Water molecules in the anion conduction pathway in the ChC are shown as sticks. c Averaged (teal mesh) chloride distribution in the transport/trimerization domain interface of GltPh in the ChC, side view. d Pore profiles of the anion pore diameter and Cl− hydration numbers, calculated as the average number of hydrogens within the first Cl− hydration shell. e Number of Cl−, Na+, and I− permeation events through ChC in simulations using either NaCl or NaI solutions at positive or negative transmembrane potentials. f Poisson–Boltzmann energies for Na+ and Cl− of WT, WTapo, and R276S GltPh in the ChC. Figure partially reprinted from [79]
The anion conduction pathway in the ChC conformation, as resolved by MD simulations, was tested with fluorescence spectroscopy and cellular electrophysiology experiments [79]. Tryptophan fluorescence is collisionally quenched by I−, and tryptophans inserted close to the simulated conduction pathway showed a significant decrease in fluorescence upon the application of I−, whereas regions remote from the simulated anion pore were not accessible to I−. As predicted from MD simulations, R276 dominates the anion selectivity of the conduction pathway and neutralization of this residue caused a loss of anion-over-cation selectivity in simulations and experiments (Fig. 5f). An in silico mutagenesis screen of all pore-forming mutants identified residues that change unitary current amplitudes and/or permit cation permeation. All substitutions that modified the anion conduction pathway in simulations affected unitary current amplitudes or Na+-to-Cl− permeation in experiments. These data demonstrate that EAATs/GltPh can adopt the anion-conducting conformation identified by MD simulations under experimental conditions. The computationally predicted channel conformation and conduction mechanism account for all published mutagenesis results on EAAT anion channels [83,84,85].
Electrophysiological experiments demonstrated that EAAT anion channel opening upon rapid substrate application is delayed compared with glutamate translocation [86]. This suggests that anion-conducting states are located outside the main transport cycle and can only be reached via branching transitions from the translocation pathway. However, crystallographic studies and single-molecule fluorescence resonance energy transfer (FRET) experiments on GltPh [87] suggested that the identified lateral movement of the transport domain might always occur during elevator translocation and, therefore, that the ChC might be visited in every transport cycle. Thus, although the molecular details of anion conduction in glutamate transporters are well understood, further functional and computational research is needed to understand the exact relationship between substrate translocation and channel opening.
HP2 is a Master Key of Glutamate Transporter Function
EAATs are amazingly complex transport proteins. They couple glutamate uptake to the symport of 3 Na+ and 1 H+, in counter transport with 1 K+, over a large range of concentrations [13, 21, 22]. They are trimers [88, 89], and each of the subunits can function independently from its neighbors [90,91,92,93]. Lastly, EAATs are not only secondary-active transporters, but also anion channels.
For almost all of these functions, HP2 has emerged as a key structural element (Fig. 6). Translocation can only occur after closure of HP2; thus, stabilization of the open or closed state by distinct combinations of bound ions and substrates ensures transport at a fixed stoichiometry. In GltPh, the internal flexibility of HP2 enables re-translocation in the apo state [67, 68] and transitions between the OFC and IFC in the absence of ligands [59]. HP2 must close to permit translocation and open to allow the effective release of substrates/ions and the association of new substrates/ions [58]. Due to its inherently flexible nature, HP2 switches between the open and closed states. Simultaneous occupation of Na1 and Na3 keeps HP2 open to permit amino acid association, which is followed by Na2 occupation and HP2 closure. Moreover, HP2 is crucial for substrate selectivity in this class of transporters. The main substrates of EAATs/GltX—l-glutamate/l-aspartate—have lower binding affinities compared with other substrates and, thus, effectively dissociate on the other membrane site. They are translocated effectively because they promote HP2 closure. This induced fit mechanism permits a combination of high selectivity and effective l-glutamate/l-aspartate transport by EAATs/GltX.
Role of HP2 in the various EAAT transport functions. Depiction of the EAAT/GltX transport cycle. From upper left: the cycle starts in the apo state for GltX and in the K+-bound state for EAATs, HP2 is closed (PDB ID: 4OYE, K+ position from [68]). In the next step, K+ is released (in EAATs) and HP2 opens, followed by the binding of two Na+ ions (PDB ID: 7AHK). Association of the substrate follows, together with the binding of one H+ in EAATs, induces HP2 closure, which permits elevator translocation (PDB ID: 2NWX, Na3 position from PDB ID: 7AHK). Translocation intermediates can open an anion-selective pore to allow thermodynamically uncoupled anion currents. From the inward closed state (PDB ID: 3KBC, Na3 position from PDB ID: 7AHK), the ligands are released upon HP2 opening. Finally, HP2 closes once again in the apo state for GltX and in the K+-bound state in EAATs to enable re-translocation (PDB ID: 3KBC, K+ position from [68])
We have wondered for decades how EAAT glutamate transporters mediate multiple transport functions, with their strict transport stoichiometry and tightly regulated anion channel gating and selectivity. Recent evidence demonstrated a surprising functional simplicity, with only few key processes responsible for this complexity. Substrates are moved across the membrane by shuttling the transport domain from the OFC to the IFC. The transport domain encompasses distinct binding sites that partially overlap and allosterically regulate HP2 opening and closing transitions, which in turn regulate access to some of these binding sites. This permits a total of six ions to be moved within a single transport cycle at fixed stoichiometry, with a clearly defined binding order, high selectivity, and highly efficient loading and unloading. In addition, this process permits the opening and closing of a selective anion channel whose physiological and pathophysiological importance we are just starting to appreciate.
Abbreviations
- ChC:
-
Channel-like conformation
- EA6:
-
Episodic ataxia 6
- EAAT:
-
Excitatory amino acid transporter
- GltPh :
-
Glutamate transporter homologue from Pyrococcus horikoshii
- GltTk :
-
Glutamate transporter homologue from Thermococcus kodakarensis
- IC:
-
Intermediate conformation
- IFC:
-
Inward-facing conformation
- MD:
-
Molecular dynamics
- OFC:
-
Outward-facing conformation
References
Danbolt NC, Pines G, Kanner BI (1990) Purification and reconstitution of the sodium- and potassium-coupled glutamate transport glycoprotein from rat brain. Biochemistry 29:6734–6740. https://doi.org/10.1021/bi00480a025
Kanner BI, Sharon I (1978) Active transport of L-glutamate by membrane vesicles isolated from rat brain. Biochemistry 17:3949–3953. https://doi.org/10.1021/bi00612a011
Kanner BI, Sharon I (1978) Solubilization and reconstitution of the L-glutamic acid transporter from rat brain. FEBS Lett 94:245–248. https://doi.org/10.1016/0014-5793(78)80947-8
Kanner BI, Marva E (1982) Efflux of l-glutamate by synaptic plasma membrane vesicles isolated from rat brain. Biochemistry 21:3143–3147. https://doi.org/10.1021/bi00256a017
Kanner BI, Bendahan A (1982) Binding order of substrates to the sodium and potassium ion coupled l-glutamic acid transporter from rat brain. Biochemistry 21:6327–6330. https://doi.org/10.1021/bi00267a044
Storck T, Schulte S, Hofmann K, Stoffel W (1992) Structure, expression, and functional analysis of a Na+-dependent glutamate/aspartate transporter from rat brain. Proc Natl Acad Sci 89:10955–10959. https://doi.org/10.1073/pnas.89.22.10955
Pines G, Danbolt NC, Bjoras M, Zhang Y, Bendahan A, Eide L, Koepsell H, Storm-Mathisen J, Seeberg E, Kanner BI (1992) Cloning and expression of a rat brain l-glutamate transporter. Nature 360:464–467. https://doi.org/10.1038/360464a0
Kanai Y, Hediger MA (1992) Primary structure and functional characterization of a high-affinity glutamate transporter. Nature 360(6403):467–471. https://doi.org/10.1038/360467a0
Arriza JL, Fairman WA, Wadiche JI, Murdoch GH, Kavanaugh MP, Amara SG (1994) Functional comparisons of three glutamate transporter subtypes cloned from human motor cortex. J Neurosci 14:5559–5569. https://doi.org/10.1523/jneurosci.14-09-05559.1994
Fairman WA, Vandenberg RJ, Arriza JL, Kavanaugh MP, Amara SG (1995) An excitatory amino-acid transporter with properties of a ligand-gated chloride channel. Nature 375:599–603. https://doi.org/10.1038/375599a0
Arriza JL, Eliasof S, Kavanaugh MP, Amara SG (1997) Excitatory amino acid transporter 5, a retinal glutamate transporter coupled to a chloride conductance. Proc Natl Acad Sci 94:4155–4160. https://doi.org/10.1073/pnas.94.8.4155
Wadiche JI, Arriza JL, Amara SG, Kavanaugh MP (1995) Kinetics of a human glutamate transporter. Neuron 14:1019–1027. https://doi.org/10.1016/0896-6273(95)90340-2
Zerangue N, Kavanaugh MP (1996) Flux coupling in a neuronal glutamate transporter. Nature 383:634–637. https://doi.org/10.1038/383634a0
Wadiche JI, Kavanaugh MP (1998) Macroscopic and microscopic properties of a cloned glutamate transporter/chloride channel. J Neurosci 18:7650–7661. https://doi.org/10.1523/jneurosci.18-19-07650.1998
Otis TS, Kavanaugh MP (2000) Isolation of current components and partial reaction cycles in the glial glutamate transporter EAAT2. J Neurosci 20:2749–2757. https://doi.org/10.1523/jneurosci.20-08-02749.2000
Wadiche JI, Amara SG, Kavanaugh MP (1995) Ion fluxes associated with excitatory amino acid transport. Neuron 15:721–728. https://doi.org/10.1016/0896-6273(95)90159-0
Gameiro A, Braams S, Rauen T, Grewer C (2011) The discovery of slowness: low-capacity transport and slow anion channel gating by the glutamate transporter EAAT5. Biophys J 100:2623–2632. https://doi.org/10.1016/j.bpj.2011.04.034
Schneider N, Cordeiro S, Machtens JP, Braams S, Rauen T, Fahlke C (2014) Functional properties of the retinal glutamate transporters GLT-1c and EAAT5. J Biol Chem 289:1815–1824. https://doi.org/10.1074/jbc.m113.517177
Larsson HP, Picaud SA, Werblin FS, Lecar H (1996) Noise analysis of the glutamate-activated current in photoreceptors. Biophys J 70:733–742. https://doi.org/10.1016/s0006-3495(96)79613-3
Fahlke C, Kortzak D, Machtens JP (2016) Molecular physiology of EAAT anion channels. Pflugers Arch 468:491–502. https://doi.org/10.1007/s00424-015-1768-3
Levy LM, Warr O, Attwell D (1998) Stoichiometry of the glial glutamate transporter GLT-1 expressed inducibly in a Chinese hamster ovary cell line selected for low endogenous Na+-dependent glutamate uptake. J Neurosci 18:9620–9628. https://doi.org/10.1523/jneurosci.18-23-09620.1998
Owe SG, Marcaggi P, Attwell D (2006) The ionic stoichiometry of the GLAST glutamate transporter in salamander retinal glia. J Physiol 577:591–599. https://doi.org/10.1113/jphysiol.2006.116830
Groeneveld M, Slotboom DJ (2010) Na+:aspartate coupling stoichiometry in the glutamate transporter homologue GltPh. Biochemistry 49:3511–3513. https://doi.org/10.1021/bi100430s
Danbolt NC (2001) Glutamate uptake. Prog Neurobiol 65:1–105. https://doi.org/10.1016/s0301-0082(00)00067-8
Petr GT, Sun Y, Frederick NM, Zhou Y, Dhamne SC, Hameed MQ, Miranda C, Bedoya EA, Fischer KD, Armsen W, Wang J, Danbolt NC, Rotenberg A, Aoki CJ, Rosenberg PA (2015) Conditional deletion of the glutamate transporter GLT-1 reveals that astrocytic GLT-1 protects against fatal epilepsy while neuronal GLT-1 contributes significantly to glutamate uptake into synaptosomes. J Neurosci 35:5187–5201. https://doi.org/10.1523/jneurosci.4255-14.2015
Rimmele TS, Rosenberg PA (2016) GLT-1: the elusive presynaptic glutamate transporter. Neurochem Int 98:19–28. https://doi.org/10.1016/j.neuint.2016.04.010
Tanaka K, Watase K, Manabe T, Yamada K, Watanabe M, Takahashi K, Iwama H, Nishikawa T, Ichihara N, Kikuchi T, Okuyama S, Kawashima N, Hori S, Takimoto M, Wada K (1997) Epilepsy and exacerbation of brain injury in mice lacking the glutamate transporter GLT-1. Science 276:1699–1702. https://doi.org/10.1126/science.276.5319.1699
Watase K, Hashimoto K, Kano M, Yamada K, Watanabe M, Inoue Y, Okuyama S, Sakagawa T, Ogawa S, Kawashima N, Hori S, Takimoto M, Wada K, Tanaka K (1998) Motor discoordination and increased susceptibility to cerebellar injury in GLAST mutant mice. Eur J Neurosci 10:976–988. https://doi.org/10.1046/j.1460-9568.1998.00108.x
Peghini P, Janzen J, Stoffel W (1997) Glutamate transporter EAAC-1-deficient mice develop dicarboxylic aminoaciduria and behavioral abnormalities but no neurodegeneration. EMBO J 16:3822–3832. https://doi.org/10.1093/emboj/16.13.3822
Perkins EM, Clarkson YL, Suminaite D, Lyndon AR, Tanaka K, Rothstein JD, Skehel PA, Wyllie DJA, Jackson M (2018) Loss of cerebellar glutamate transporters EAAT4 and GLAST differentially affects the spontaneous firing pattern and survival of Purkinje cells. Hum Mol Genet 27:2614–2627. https://doi.org/10.1093/hmg/ddy169
Veruki ML, Morkve SH, Hartveit E (2006) Activation of a presynaptic glutamate transporter regulates synaptic transmission through electrical signaling. Nat Neurosci 9:1388–1396. https://doi.org/10.1038/nn1793
Wersinger E, Schwab Y, Sahel JA, Rendon A, Pow DV, Picaud S, Roux MJ (2006) The glutamate transporter EAAT5 works as a presynaptic receptor in mouse rod bipolar cells. J Physiol 577:221–234. https://doi.org/10.1113/jphysiol.2006.118281
Jen JC, Wan J, Palos TP, Howard BD, Baloh RW (2005) Mutation in the glutamate transporter EAAT1 causes episodic ataxia, hemiplegia, and seizures. Neurology 65:529–534. https://doi.org/10.1212/01.wnl.0000172638.58172.5a
Winter N, Kovermann P, Fahlke C (2012) A point mutation associated with episodic ataxia 6 increases glutamate transporter anion currents. Brain 135:3416–3425. https://doi.org/10.1093/brain/aws255
Hotzy J, Schneider N, Kovermann P, Fahlke C (2013) Mutating a conserved proline residue within the trimerization domain modifies Na+ binding to excitatory amino acid transporters and associated conformational changes. J Biol Chem 288:36492–36501. https://doi.org/10.1074/jbc.m113.489385
Kovermann P, Untiet V, Kolobkova Y, Engels M, Baader S, Schilling K, Fahlke C (2020) Increased glutamate transporter-associated anion currents cause glial apoptosis in episodic ataxia 6. Brain Commun 2:fcaa022. https://doi.org/10.1093/braincomms/fcaa022
Untiet V, Kovermann P, Gerkau NJ, Gensch T, Rose CR, Fahlke C (2017) Glutamate transporter-associated anion channels adjust intracellular chloride concentrations during glial maturation. Glia 65:388–400. https://doi.org/10.1002/glia.23098
Chivukula AS, Suslova M, Kortzak D, Kovermann P, Fahlke C (2020) Functional consequences of SLC1A3 mutations associated with episodic ataxia 6. Hum Mutat 41:1892–1905. https://doi.org/10.1002/humu.24089
Kovermann P, Hessel M, Kortzak D, Jen JC, Koch J, Fahlke C, Freilinger T (2017) Impaired K+ binding to glial glutamate transporter EAAT1 in migraine. Sci Rep 7:13913. https://doi.org/10.1038/s41598-017-14176-4
Adamczyk A, Gause CD, Sattler R, Vidensky S, Rothstein JD, Singer H, Wang T (2011) Genetic and functional studies of a missense variant in a glutamate transporter, SLC1A3, in Tourette syndrome. Psychiatr Genet 21:90–97. https://doi.org/10.1097/ypg.0b013e328341a307
van Amen-Hellebrekers CJ, Jansen S, Pfundt R, Schuurs-Hoeijmakers JH, Koolen DA, Marcelis CL, de Leeuw N, de Vries BB (2016) Duplications of SLC1A3: associated with ADHD and autism. Eur J Med Genet 59:373–376. https://doi.org/10.1016/j.ejmg.2016.06.003
Guella I, McKenzie MB, Evans DM, Buerki SE, Toyota EB, Van Allen MI, Epilepsy Genomics Study, Suri M, Elmslie F, Deciphering Developmental Disorders Study, Simon MEH, van Gassen KLI, Héron D, Keren B, Nava C, Connolly MB, Demos M, Farrer MJ (2017) De Novo mutations in YWHAG cause early-onset epilepsy. Am J Hum Genet 101:300–310. https://doi.org/10.1016/j.ajhg.2017.07.004
Stergachis AB, Pujol-Giménez J, Gyimesi G, Fuster D, Albano G, Troxler M, Picker J, Rosenberg PA, Bergin A, Peters J, El Achkar CM, Harini C, Manzi S, Rotenberg A, Hediger MA, Rodan LH (2019) Recurrent SLC1A2 variants cause epilepsy via a dominant negative mechanism. Ann Neurol 85:921–926. https://doi.org/10.1002/ana.25477
Bailey CG, Ryan RM, Thoeng AD, Ng C, King K, Vanslambrouck JM, Auray-Blais C, Vandenberg RJ, Broer S, Rasko JE (2011) Loss-of-function mutations in the glutamate transporter SLC1A1 cause human dicarboxylic aminoaciduria. J Clin Investig 121:446–453. https://doi.org/10.1172/jci44474
Yernool D, Boudker O, Jin Y, Gouaux E (2004) Structure of a glutamate transporter homologue from Pyrococcus horikoshii. Nature 431:811–818. https://doi.org/10.1038/nature03018
Boudker O, Ryan RM, Yernool D, Shimamoto K, Gouaux E (2007) Coupling substrate and ion binding to extracellular gate of a sodium-dependent aspartate transporter. Nature 445:387–393. https://doi.org/10.1038/nature05455
Jensen S, Guskov A, Rempel S, Hanelt I, Slotboom DJ (2013) Crystal structure of a substrate-free aspartate transporter. Nat Struct Mol Biol 20:1224–1226. https://doi.org/10.1038/nsmb.2663
Arkhipova V, Guskov A, Slotboom DJ (2020) Structural ensemble of a glutamate transporter homologue in lipid nanodisc environment. Nat Commun 11:998. https://doi.org/10.1038/s41467-020-14834-8
Alleva C, Kovalev K, Astashkin R, Berndt M, Baeken C, Balandin T, Gordeliy V, Fahlke C, Machtens JP (2020) Na+-dependent gate dynamics and electrostatic attraction ensure substrate coupling in glutamate transporters. Sci Adv 6:eaba9854. https://doi.org/10.1126/sciadv.aba9854
Reyes N, Ginter C, Boudker O (2009) Transport mechanism of a bacterial homologue of glutamate transporters. Nature 462:880–885. https://doi.org/10.1038/nature08616
Verdon G, Boudker O (2012) Crystal structure of an asymmetric trimer of a bacterial glutamate transporter homolog. Nat Struct Mol Biol 19:355–357. https://doi.org/10.1038/nsmb.2233
Guskov A, Jensen S, Faustino I, Marrink SJ, Slotboom DJ (2016) Coupled binding mechanism of three sodium ions and aspartate in the glutamate transporter homologue GltTk. Nat Commun 7:13420. https://doi.org/10.1038/ncomms13420
Crisman TJ, Qu S, Kanner BI, Forrest LR (2009) Inward-facing conformation of glutamate transporters as revealed by their inverted-topology structural repeats. Proc Natl Acad Sci 106:20752–20757. https://doi.org/10.1073/pnas.0908570106
Canul-Tec JC, Assal R, Cirri E, Legrand P, Brier S, Chamot-Rooke J, Reyes N (2017) Structure and allosteric inhibition of excitatory amino acid transporter 1. Nature 544:446–451. https://doi.org/10.1038/nature22064
Garaeva AA, Guskov A, Slotboom DJ, Paulino C (2019) A one-gate elevator mechanism for the human neutral amino acid transporter ASCT2. Nat Commun 10:3427. https://doi.org/10.1038/s41467-019-11363-x
Yu X, Plotnikova O, Bonin PD, Subashi TA, McLellan TJ, Dumlao D, Che Y, Dong YY, Carpenter EP, West GM, Qiu X, Culp JS, Han S (2019) Cryo-EM structures of the human glutamine transporter SLC1A5 (ASCT2) in the outward-facing conformation. eLife 8:E48120. https://doi.org/10.7554/elife.48120
Garaeva AA, Oostergetel GT, Gati C, Guskov A, Paulino C, Slotboom DJ (2018) Cryo-EM structure of the human neutral amino acid transporter ASCT2. Nat Struct Mol Biol 25:515–521. https://doi.org/10.1038/s41594-018-0076-y
Verdon G, Oh S, Serio RN, Boudker O (2014) Coupled ion binding and structural transitions along the transport cycle of glutamate transporters. eLife 3:e02283. https://doi.org/10.7554/elife.02283
Matin TR, Heath GR, Huysmans GHM, Boudker O, Scheuring S (2020) Millisecond dynamics of an unlabeled amino acid transporter. Nat Commun 11:5016. https://doi.org/10.1038/s41467-020-18811-z
Bastug T, Heinzelmann G, Kuyucak S, Salim M, Vandenberg RJ, Ryan RM (2012) Position of the third Na+ site in the aspartate transporter GltPh and the human glutamate transporter, EAAT1. PLoS ONE 7:e33058. https://doi.org/10.1371/journal.pone.0033058
Hänelt I, Jensen S, Wunnicke D, Slotboom DJ (2015) Low affinity and slow Na+ binding precedes high affinity aspartate binding in the secondary-active transporter GltPh. J Biol Chem 290:15962–15972. https://doi.org/10.1074/jbc.m115.656876
Focke PJ, Moenne-Loccoz P, Larsson HP (2011) Opposite movement of the external gate of a glutamate transporter homolog upon binding cotransported sodium compared with substrate. J Neurosci 31:6255. https://doi.org/10.1523/jneurosci.6096-10.2011
Chakraborty P, Di Cera E (2017) Induced fit is a special case of conformational selection. Biochemistry 56:2853–2859. https://doi.org/10.1021/acs.biochem.7b00340
Ewers D, Becher T, Machtens JP, Weyand I, Fahlke C (2013) Induced fit substrate binding to an archeal glutamate transporter homologue. Proc Natl Acad Sci 110:12486–12491. https://doi.org/10.1073/pnas.1300772110
Oh S, Boudker O (2018) Kinetic mechanism of coupled binding in sodium-aspartate symporter GltPh. eLife 7:e37291. https://doi.org/10.7554/eLife.37291.001
Gaillard I, Slotboom DJ, Knol J, Lolkema JS, Konings WN (1996) Purification and reconstitution of the glutamate carrier GltT of the thermophilic bacterium Bacillus stearothermophilus. Biochemistry 35:6150–6156. https://doi.org/10.1021/bi953005v
Ryan RM, Compton EL, Mindell JA (2009) Functional characterization of a Na+-dependent aspartate transporter from Pyrococcus horikoshii. J Biol Chem 284:17540–17548. https://doi.org/10.1074/jbc.m109.005926
Kortzak D, Alleva C, Weyand I, Ewers D, Zimmermann MI, Franzen A, Machtens JP, Fahlke C (2019) Allosteric gate modulation confers K+ coupling in glutamate transporters. EMBO J 38:e101468. https://doi.org/10.15252/embj.2019101468
Wang J, Zhang K, Goyal P, Grewer C (2020) Mechanism and potential sites of potassium interaction with glutamate transporters. J Gen Physiol 152:e202012577. https://doi.org/10.1085/jgp.202012577
Grewer C, Zhang Z, Mwaura J, Albers T, Schwartz A, Gameiro A (2012) Charge compensation mechanism of a Na+-coupled, secondary active glutamate transporter. J Biol Chem 287:26921–26931. https://doi.org/10.1074/jbc.m112.364059
LeVine MV, Cuendet MA, Khelashvili G, Weinstein H (2016) Allosteric mechanisms of molecular machines at the membrane: transport by sodium-coupled symporters. Chem Rev 116:6552–6587. https://doi.org/10.1021/acs.chemrev.5b00627
Forrest LR, Tavoulari S, Zhang YW, Rudnick G, Honig B (2007) Identification of a chloride ion binding site in Na+/Cl–dependent transporters. Proc Natl Acad Sci 104:12761–12766. https://doi.org/10.1073/pnas.0705600104
Zomot E, Bendahan A, Quick M, Zhao Y, Javitch JA, Kanner BI (2007) Mechanism of chloride interaction with neurotransmitter:sodium symporters. Nature 449:726–730. https://doi.org/10.1038/nature06133
Zerangue N, Kavanaugh MP (1996) ASCT-1 is a neutral amino acid exchanger with chloride channel activity. J Biol Chem 271:27991–27994. https://doi.org/10.1074/jbc.271.45.27991
Sonders MS, Amara SG (1996) Channels in transporters. Curr Opin Neurobiol 6:294–302. https://doi.org/10.1016/s0959-4388(96)80111-5
Otis TS, Jahr CE (1998) Anion currents and predicted glutamate flux through a neuronal glutamate transporter. J Neurosci 18:7099–7110. https://doi.org/10.1523/jneurosci.18-18-07099.1998
Bergles DE, Tzingounis AV, Jahr CE (2002) Comparison of coupled and uncoupled currents during glutamate uptake by GLT-1 transporters. J Neurosci 22:10153–10162. https://doi.org/10.1523/jneurosci.22-23-10153.2002
Machtens JP, Kovermann P, Fahlke C (2011) Substrate-dependent gating of anion channels associated with excitatory amino acid transporter 4. J Biol Chem 286:23780–23788. https://doi.org/10.1074/jbc.m110.207514
Machtens JP, Kortzak D, Lansche C, Leinenweber A, Kilian P, Begemann B, Zachariae U, Ewers D, de Groot BL, Briones R, Fahlke C (2015) Mechanisms of anion conduction by coupled glutamate transporters. Cell 160:542–553. https://doi.org/10.1016/j.cell.2014.12.035
Melzer N, Biela A, Fahlke C (2003) Glutamate modifies ion conduction and voltage-dependent gating of excitatory amino acid transporter-associated anion channels. J Biol Chem 278:50112–50119. https://doi.org/10.1074/jbc.m307990200
Kovermann P, Machtens JP, Ewers D, Fahlke C (2010) A conserved aspartate determines pore properties of anion channels associated with excitatory amino acid transporter 4 (EAAT4). J Biol Chem 285:23676–23686. https://doi.org/10.1074/jbc.m110.126557
Machtens JP, Fahlke C, Kovermann P (2011) Noise analysis to study unitary properties of transporter-associated ion channels. Channels 5:468–474. https://doi.org/10.4161/chan.5.6.17453
Shabaneh M, Rosental N, Kanner BI (2014) Disulfide cross-linking of transport and trimerization domains of a neuronal glutamate transporter restricts the role of the substrate to the gating of the anion conductance. J Biol Chem 289:11175–11182. https://doi.org/10.1074/jbc.m114.550277
Cater RJ, Vandenberg RJ, Ryan RM (2016) Tuning the ion selectivity of glutamate transporter-associated uncoupled conductances. J Gen Physiol 148:13–24. https://doi.org/10.1085/jgp.201511556
Cheng MH, Torres-Salazar D, Gonzalez-Suarez AD, Amara SG, Bahar I (2017) Substrate transport and anion permeation proceed through distinct pathways in glutamate transporters. eLife 6:e25850. https://doi.org/10.7554/elife.25850
Grewer C, Watzke N, Wiessner M, Rauen T (2000) Glutamate translocation of the neuronal glutamate transporter EAAC1 occurs within milliseconds. Proc Natl Acad Sci 97:9706–9711. https://doi.org/10.1073/pnas.160170397
Akyuz N, Georgieva ER, Zhou Z, Stolzenberg S, Cuendet MA, Khelashvili G, Altman RB, Terry DS, Freed JH, Weinstein H, Boudker O, Blanchard SC (2015) Transport domain unlocking sets the uptake rate of an aspartate transporter. Nature 518:68–73. https://doi.org/10.1038/nature14158
Gendreau S, Voswinkel S, Torres-Salazar D, Lang N, Heidtmann H, Detro-Dassen S, Schmalzing G, Hidalgo P, Fahlke C (2004) A trimeric quaternary structure is conserved in bacterial and human glutamate transporters. J Biol Chem 279:39505–39512. https://doi.org/10.1074/jbc.m408038200
Nothmann D, Leinenweber A, Torres-Salazar D, Kovermann P, Hotzy J, Gameiro A, Grewer C, Fahlke C (2011) Hetero-oligomerization of neuronal glutamate transporters. J Biol Chem 286:3935–3943. https://doi.org/10.1074/jbc.m110.187492
Grewer C, Balani P, Weidenfeller C, Bartusel T, Tao Z, Rauen T (2005) Individual subunits of the glutamate transporter EAAC1 homotrimer function independently of each other. Biochemistry 44:11913–11923. https://doi.org/10.1021/bi050987n
Leary GP, Stone EF, Holley DC, Kavanaugh MP (2007) The glutamate and chloride permeation pathways are colocalized in individual neuronal glutamate transporter subunits. J Neurosci 27:2938–2942. https://doi.org/10.1523/jneurosci.4851-06.2007
Koch HP, Brown RL, Larsson HP (2007) The glutamate-activated anion conductance in excitatory amino acid transporters is gated independently by the individual subunits. J Neurosci 27:2943–2947. https://doi.org/10.1523/jneurosci.0118-07.2007
Erkens GB, Hänelt I, Goudsmits JM, Slotboom DJ, van Oijen AM (2013) Unsynchronised subunit motion in single trimeric sodium-coupled aspartate transporters. Nature 502:119–123. https://doi.org/10.1038/nature12538
Funding
Open Access funding enabled and organized by Projekt DEAL. This work was supported by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) to Ch.F. (FA 301/12-2) and J.P.M (MA 7525/1-2) as part of the Research Unit FOR 2518, DynIon, project P4. The authors gratefully acknowledge the computing time granted through JARA on the supercomputer JURECA at Forschungszentrum Jülich and the supercomputer CLAIX at RWTH Aachen University.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare no competing financial interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Special Issue: In Honor of Prof. Baruch Kanner.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Alleva, C., Machtens, JP., Kortzak, D. et al. Molecular Basis of Coupled Transport and Anion Conduction in Excitatory Amino Acid Transporters. Neurochem Res 47, 9–22 (2022). https://doi.org/10.1007/s11064-021-03252-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11064-021-03252-x