Abstract
Actin is a cytoskeletal protein which forms dense, highly interconnected networks within eukaryotic cells. A growing body of evidence suggests that actin-mediated intra- and extracellular signalling is instrumental in facilitating organism-level emergent behaviour patterns which, crucially, may be characterised as natural expressions of computation. We use excitable cellular automata modelling to simulate signal transmission through cell arrays whose topology was extracted from images of Watershed transformation-derived actin network reconstructions; the actin networks sampled were from laboratory experimental observations of a model organism, slime mould Physarum polycephalum. Our results indicate that actin networks support directional transmission of generalised energetic phenomena, the amplification and trans-network speed of which of which is proportional to network density (whose primary determinant is the anatomical location of the network sampled). Furthermore, this model also suggests the ability of such networks for supporting signal-signal interactions which may be characterised as Boolean logical operations, thus indicating that a cell’s actin network may function as a nanoscale data transmission and processing network. We conclude by discussing the role of the cytoskeleton in facilitating intracellular computing, how computation can be implemented in such a network and practical considerations for designing ‘useful’ actin circuitry.
Similar content being viewed by others
1 Introduction
The cytoskeleton is a ubiquitous organelle in eukaryotic cells that comprises a scaffold of proteins whose topology dynamically rearranges as the cell migrates or deforms under mechanical pressure. Where it was once thought that the primary function of the cytoskeleton was to provide mechanical support to the cell, it is now clear that it also participates in a multitude of intracellular processes, including trafficking of organelles through the cytoplasm, facilitating cell movement via participation in muscular contraction and transduction of energetic/signalling events through the cell (Janmey 1998). Although there are a great many varieties of protein that may be present in a cell’s cytoskeleton, the predominant two are tubulin, which forms long tubular structures (microtubules), and actin, which forms smaller filamentous double helices (microfilaments). We here focus on modelling signal transmission through networks composed of the latter protein, through the use of excitable cellular automata (CA).
The transmission of mechanical force through actin microfilaments coupled to cell surface mechanoreceptors is one of the more thoroughly characterised forms of energetic event which the cytoskeleton is able to conduct (Janmey 1998), but there is a growing body of evidence to suggest that further forms of intracellular signal are carried by the cytoskeleton such as ionic waves, vesicle-bound signalling molecules and quantum events such as solitons (Carpenter 2000; Craddock et al. 2012; Davydov 1977; Forgacs et al. 2004; Maniotis et al. 1997; Schmidt and Hall 1998; Tuszyński et al. 2004; Mayne and Adamatzky 2015). It has been suggested by several authors that many of the emergent properties displayed by organisms (e.g. synergistic cooperation between cells in complex organ systems such as the brain) arise at the sub-cellular level from non-linear interactions of the myriad signalling processes mediated by the cytoskeleton. If this is indeed the case, then by extension any research furthering our knowledge of cytoskeleton-mediated processes will enhance our understanding of poorly-characterised phenomena such as brain function and hence precipitate a new wave of technologies and medical therapies (Hameroff 1987; Priel et al. 2010).
From the perspective of computer science, the advancement of this topic is enthusing as biological signalling processes can, when interpreted in the language of computation, represent intracellular ‘data’ that are the consequences of environmental ‘input’ from cell-surface reception. Unstructured sensory data is, when transmitted by the cytoskeleton, transduced into a regular, repeatable format that the cell is able to interpret in an unambiguous manner.
Whilst we are not the first authors to comment on the putatively computational nature of energetic cytoskeletal processes, the majority of the work to date on cytoskeletal computing focuses on microtubules and typically concerns modelling the cytoskeleton as a conventional general purpose computer. It has been previously demonstrated, for example, that microtubules may transmit propagating waves of protein conformational state transitions and hence may be thought of as a data bus; Boolean logical operations have been suggested to occur when these events interact with the microtubule-associated proteins that link microtubules to other cytoskeletal components (Craddock et al. 2012; Lahoz-Beltra et al. 1993). We emphasise, conversely, that the functionality of a cell is, whilst analogous to a conventional computer in some aspects, so divergent from silicon-based architectures that any direct comparisons between the two are null; consequently, practical computing devices based on biological substrates must be built on emphatically unconventional paradigms.
The work presented here concerns using CA to simulate generalised information transmission events through experimentally-derived cytoskeletal network conformations towards indicating the characteristics of the organelle as a computing resource. We conclude by discussing practical aspects of cytoskeletal circuit design. The laboratory experimental aspects of this study involved sampling actin networks from a model organism, slime mould Physarum polycephalum (Fig. 1); slime mould is an unconventional computing substrate in its own right (we refer the reader to Refs. Adamatzky (2010) for an overview of slime mould computing and Mayne et al. (2015a) for a review of the concept of cytoskeletal computing within slime mould), our use of this organism in this context is simply as a malleable eukaryotic cell capable of expressing a range of emergent behaviours and possessing an abundant cytoskeletal network.
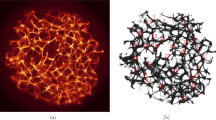
The P. polycephalum cytoskeleton is not as well characterised as the mammalian equivalent, but the constituent proteins are highly conserved between species: slime mould microfilaments, as with their mammalian counterparts, are composed of globular actin monomers (g-actin) whose quarternary structure is a double helix, individual fibres of which are known as f-actin (Fig. 2). The plasmodial actin network spans the entire cell but is most concentrated in a dense cortical region about the cell’s periphery (the ectoplasm), where it articulates onto membrane-bound proteins. The P. polycephalum advancing anterior margin (Fig. 1), which is formed from the confluence of multiple pseudopodia, contains denser actin networks than its caudal tubular regions (Alexopoulos 1982; Mayne et al. 2015a). Whilst pseudopodium formation via momentary assembly of actin is a well-observed phenomenon, this does pose the question of how such topological dimorphism impacts on intracellular communicative processes, hence investigation into this was made a secondary goal of this study.
Schematic diagram of an actin microfilament as a double helix structure composed of individual G-actin monomers. Scale bar approx. 5 nm, helix twist not to scale. Adapted with permission from Adamatzky and Mayne (2015)
2 Methods
2.1 Physarum polycephalum culture and microscopy
Stock plasmodia of P. polycephalum (strain HU554 \(\times\) HU560) were cultivated on 2 % non-nutrient agar (NNA) plates at room temperature in the absence of light. They were provided with porridge oats as a nutrient substrate and were sub-cultured routinely every 3–4 days, as required.
Actin was labelled for confocal microscopy as follows: two hemispheres of NNA were prepared on large glass microscope coverslips by dripping \(2\times 0.5\) ml of molten agar onto them with a pipette, with a gap of approximately 10 mm separating the two. Samples of stock plasmodium were homogenised with a scalpel blade and transferred to one NNA hemisphere. An oat flake was placed on the second hemisphere to encourage the organism to propagate between them; the cover slip was placed in an air-tight Petri dish, which was left in the dark at room temperature. Samples of the anterior margin were taken by chemically fixing the organism before its advancing regions had finished propagating across the gap between the hemispheres and were hence spanning the naked glass, whereas plasmodial tubes were prepared by waiting until the second hemisphere had been fully colonised, as this typically resulted in the gap between the two hemispheres being linked by a single large tube.
Fixation was achieved by flooding the Petri dish with 2 % paraformaldehyde in pH 7.2 phosphate-buffered saline solution for 1 h. This was followed by 3 × 5 min rinses in the same buffer, after which the sample was permeabalised with 0.1 % Triton X-100. Following further rinsing and draining, Alexa Fluor 488 Phalloidin (Molecular Probes, USA) was added for 1 h at a concentration of 5 μg/ml in methanol. The samples were then washed again before being stained with DAPI (Abcam, UK).
Confocal microscopy was performed with a Perkin Elmer UltraView ERS FRET-H spinning disk confocal laser scanning microscope.Footnote 1
2.2 Network extraction and CA model
Actin networks were reconstructed using the Watershed transformation on greyscale confocal micrographs with Matlab 2015a (Mathworks, USA). To represent the topology of actin network reconstructions in 2D cellular automaton models we assigned cells corresponding to each black pixel as conductive; other cells are assigned to be non-conductive. Although any reconstruction of nanoscale structures using a light microscopic technique necessitates a certain degree of inaccuracy, the watershed transformation was chosen for it having previously been demonstrated as a viable technique for approximating actin network topologies to a suitable degree of accuracy (Fleischer et al. 2007). The concave hull of extracted networks was also computed in instances where the cell’s membrane was present in order to demarcate the cell’s boundaries.
An excitable CA representing the propagation of generalised energetic events along actin microfilaments was constructed as follows. In an excitable medium, discretised spaces (cells) are considered to be in one of three states (Greenberg and Hastings 1978):
-
1.
Resting A stable equilibrium state.
-
2.
Excited The energy level of the cell is temporarily higher than the equilibrium level; in context with this investigation, this could correspond to a breather propagating through a particular actin molecule or a local electrochemical disturbance caused by ionic wave trasmission through a microfilament.
-
3.
Refractory A temporary state directly following excitation wherein the cell is in the process of returning to equilibrium and cannot be re-excited until it has re-entered a resting state.
Thus in an excitable CA, the excitable medium is represented as a 2D array of cells, wherein each cell takes one of these three states. The array was initialised by importing 2D actin network reconstructions as 500 × 500 pixel bitmap images wherein each pixel represented a cell: as only black-coloured cells (corresponding to the presence of actin in these pixels in the original image) update their states and other cells remain resting indefinitely, the excitation may only propagate through the reconstructed network. A resting black cell becomes excited (indicated by turning pink in the simulation, see videos 1 and 2 in Electronic Supplementary Material) during time iteration \(t+1\) if it has at least one excited neighbouring cell (von Neumann neighbourhood) at time t, thus ‘signals’—represented by excitation states—can be observed to propagate through the reconstructed networks over time. Cells remain in an excited state for one time iteration before turning into a refractory cell (denoted by turning blue in the simulation, see videos in Electronic Supplementary Material), also for one time iteration, after which it reverts into a resting cell. When a wave of excitation, propagating along conductive cells, ‘collides’ with another excitation wave head-on, both signals annihilate as their being flanked by refractory cells disallows their moving ‘through’ or ricocheting from each other. Excitations were initiated at random locations on the periphery of reconstructed networks to represent the initiation of an energetic event, e.g. through stimulation of a cell surface receptor.
Integral excitation dynamics of extracted networks were measured by computing the number of cells becoming excited each time iteration. The comparative speed of transmission through these networks was assessed by measuring the number of timesteps taken for the signal to reach a point in the network an arbitrary but fixed distance away: \(450\pm 1.2\) away in all experiments.
All experiments (visualisation of an area, running of CA and measurement of excitation dynamics) were repeated 5 times for each anatomical region sampled.
3 Results
3.1 Visualisation of actin
In corroboration with previously published findings, the plasmodial actin network was observed to be concentrated in the cell’s cortical regions and appeared to form associations with the surface of the organism. Actin networks appeared denser in the organism’s anterior margin (Fig. 3a) than in plasmodial tubes (Fig. 4a).
3.2 Modelling information flow through extracted actin networks
Exemplar extracted actin networks are shown in Figs. 3b and 4b; video footage of the CA model being run on both of these networks is included as supplementary information (see Electronic Supplementary Material). In both, the initial perturbation was observed to propagate through the entire network thanks to the signal becoming amplified when it met network junctions, as can be observed through the gradual increase in number of excited actin units over time in Figs 3c and 4c. Excitation dynamics were found to be richer in the anterior regions of the plasmodium. Network topologies were found to be structured such that signal back-propagation rarely occurred; when it did occur, reversed signals usually collided with signals travelling in the opposite direction and annihilated.
The speed (in timesteps) for a signal to travel to a network location 450 pixels away was found to be faster in anterior margin networks in every instance (659 and 829 time steps in the two examples given, respectively).
Actin excitation dynamics in the P. polycephalum plasmodial tube. a Confocal micrograph. b Extracted network. Excitation point indicated by arrow. c Integral excitation dynamics of actin network, which are less rich than in Fig. 3c; trendline is a polynomial fit. a–bScale bar 50 μm
4 Discussion
4.1 On dynamic actin network transformations
Actin network density was found to be inversely proportional to proximity to active growth areas in the organism’s anterior regions; this observation was substantiated by a richer excitation dynamics and greater speed of signal transduction through reconstructed anterior margin networks using excitable CA. Thus, we can speculate that depending on momentary actual demands of the slime mould’s physiology, the topology of its communication network will alter in density and connectivity.
It would seem that a greater degree of signal amplification and transduction speed are desirable characteristics for the protein networks of active growth regions in order to maximise environmental sensing and the organism’s ability to consequently react to input data. These characteristics are presumably reduced in caudal regions of the organism in order to preserve energy where they are not required, although we concede that other functions of the actin cytoskeleton that have not been examined here will also play a role in determining network topology. Biological substrates are often quoted as being ‘massively parallel’ in their ability to concurrently sense input from an almost incalculably large number of sensory data streams (Stepney et al. 2005); our findings emphasise the efficiency with which such substrates may optimise these processes in response to evolutionary selection pressures, thus highlighting their value to the field of unconventional computing and bio-inspired computing design.
4.2 Sensoriactuation networks for intracellular computing
Let us consider how intracellular computation may be influenced by data network topology. Computing with algorithms based on planar graphs have been demonstrated to be viable for implementing general-purpose computation: furthermore, certain classes of device —Kolmogorov-Uspensky Machines (Kolmogorov and Uspensky 1958)—whose physical architecture are based on the retrieval of data stored in dynamically-rearranging undirected graph structures, demonstrate the viability of computers built upon principles of motile components. We may speculate, therefore, that analogous processes are integrated via actin-based cellular communications which provoke morphological rearrangement. To delineate, consider the following example: despite lacking a brain or nervous tissue, a slime mould is able to ‘decide’ which of the myriad chemical gradients one of its advancing margins senses to follow. The cell’s cortical actin network becomes innervated via the activation of second messenger pathways incident of chemoreceptor stimulation; by the majority vote, the sites of the organism with the greatest amount of chemoreception (corresponding to multiple receptors being activated) are the sites of consequent directional tip growth and thus the network rearranges in the process of migration.
This is an interesting perspective as it implies that the parallelism inherent in biological computing substrates occurs as a function of the materials they are composed from and does not necessitate expending energy on the coordination or synchronisation of these behaviours. As such, the complexity of the organism is reduced whilst enabling the spontaneous generation of complex behaviours—in effect, this is a definition of emergence. These observations are complimentary to recent advances in the field of morphological computation and entity embodiment which posit that ‘outsourcing’ a certain amount of computational work to the morphology of data streams is essential in the design of artificially intelligent entities (Hauser et al. 2012; Lungarella and Sporns 2006). This concept is intimately linked to data ‘structuring’—transducing and transmitting signals in a repeatable and unambiguous manner—which is another important concept in morphological computation and control theory (Füchslin et al. 2013; Hauser et al. 2012). By logical extension, this would seem to suggest that the cytoskeleton is a medium for structuring sensorimotor data streams and hence that the emergent behaviours displayed by cells may be a product of cytoskeletal processes.
In the model presented, back-propagation of signals appeared to be curtailed via destructive signal–signal interactions—in effect, this describes collision-based computation (Adamatzky 2002). Combined with the incidence of signal amplification, we may characterise these phenomena as the following operations:
-
fan-out; signal amplification when a signal reaches a network bifurcation.
-
xor; when two signals converge following a branch, they may interact destructively such that the output configuration of the branch point is \(\langle A\cdot \bar{B} + \bar{A} \cdot B \rangle\).
Thus we may speculate that this computation-like process plays a crucial role in regulating signal propagation through the organism’s actin network.
To address a pertinent criticism of the work presented here, the modelled excitation dynamics presented are based on static, two-dimensional characterisations of dynamic, three-dimensional structures superimposed onto a two-dimensional image: as such, results presented here are purely illustrative. We note, however, that these techniques may be applied to three- or even four-dimensional image stacks; this will be a focus for future research.
4.3 Practical actin computing
Actin, along with its companion motor protein myosin, is instrumental in the retention and transport of vesicle-bound biomolecules and minerals (DePina and Langford 1999); we have previously described the movements of calcium-filled vesicles travelling down P. polycephalum actin fibres as an example of naturally-occurring collision-based computing (Mayne and Adamatzky 2015). Following the results described here, the same may also be said for other energetic events, such as quantum phenomena or/and electrical potential propagated in the form of ionic waves. ‘Hijacking’ these processes is a viable route towards the generation of practical biological computing architectures.
To engineer practical actin computing circuits we are presented with a few technical considerations. First is to generate an actin network in a desired topology; this may be in vitro or in vivo. Although minute manipulation of actin fibres has been demonstrated in vitro (Lin and Cantiello 1993), we suggest that actin growth within model organisms such as P. polycephalum is also a viable method to this end as growth patterns are essentially programmable by manipulation of the organism’s environment and internal feedback mechanisms.
The second consideration is the initiation and synchronisation of informational events. Again, this issue may best be addressed by the manipulation of environmental and internal factors, but may also be induced artificially. In slime mould models, for example, actin network contraction can be initiated by tactile stimulation (Mayne et al. 2015). The interaction environment’s topology plays an important role in synchronisation, especially in collision-based models as factors such as filament length and delay elements are necessarily functions of network topology.
4.4 Conclusion
A prime motivation into researching biological computing substrates such as slime mould is their amorphism—a property that implies architecture-less massive parallel processing. Our results indicate that the emergent behavioural capabilities of a live cell may be a product of its cytoskeletal topology and the information processing events therein. This implies that emergent behaviour is a product of the physical properties of the data network; this has important implications for our understanding of concepts such as human intelligence, which is commonly regarded as a product (in part, at least) of neural network morphology and the characteristics of the junctions (synapses) between them. In future works, we will expand this concept with experimental implementations of actin-based computing.
Notes
Image Processing Photographs were taken with a Samsung I9300 digital camera. Confocal micrographs were digitally post-processed in Volocity (Improvision, UK), and received colour assignment, deconvolution and brightness/colour adjustments. Unprocessed image files will be made available on request.
References
Adamatzky A (2002) Collision-based computing. Springer, Berlin
Adamatzky A (2010) Physarum machines: computers from slime mould. World Scientific Publishing, London
Adamatzky A, Mayne R (2015) Actin automata: phenomenology and localizations. Int J Bifurc Chaos 25(2):1550030
Alexopoulos C (1982) The biology of physarum and didymium. Cell biology: a series of monographs, chapter 5: plasmodial structure and motility. Academic Press, Cambridge
Carpenter C (2000) Actin cytoskeleton and cell signalling. Crit Care Med 28(4):94–99
Craddock T, Tuszyński J, Hameroff S (2012) Cytoskeletal signalling: is memory encoded in microtubule lattices by camkii phosphorylation? PLoS Comput Biol 8(3):e1002421
Davydov A (1977) Solitons and energy transfer along protein molecules. J Theor Biol 66:379–387
DePina A, Langford G (1999) Vesicle transport: the role of actin filaments and myosin motors. Microsc Res Tech 47(2):93–106
Fleischer F, Ananthakrishnan R, Eckel S, Schmidt H, Käs J, Svitkina T, Schmidt V, Beil M (2007) Actin network architecture and elasticity in lamellipodia of melanoma cells. New J Phys 9(11):420
Forgacs G, Yook SH, Janmey P, Jeong H, Burd C (2004) Role of the cytoskeleton in signalling networks. J Cell Sci 117(3):2769–2775
Füchslin R, Dzyakanchuk A, Flumini D, Hauser H, Hunt K, Luchsinger R, Reller B, Scheidegger S, Walker R (2013) Morphological computation and morphological control: steps towards a formal theory and applications. Artif Life 19:9–34
Greenberg J, Hastings S (1978) Spatial patterns for discrete models of diffusion in excitable media. SIAM J Appl Math 34(3):515–523
Hameroff S (1987) Ultimate computing: biomolecular consciousness and nanotechnology. Elsevier, Amsterdam
Hauser H, Ijspeert AJ, Füchslin RM, Pfeifer R, Maass W (2012) Towards a theoretic foundation for morphological computation with compliant bodies. Biol Cybern 105:355–370
Janmey P (1998) The cytoskeleton and cell signalling: component localization and mechanical coupling. Phys Rev 78(3):763–781
Kolmogorov A, Uspensky V (1958) On the definition of an algorithm. Uspekhi Matematicheskikh Nauk 13(4):3–28
Lahoz-Beltra R, Hameroff S, Dayhoff J (1993) Cytoskeletal logic: a model for molecular computation via boolean operations in microtubules and microtubule-associated proteins. Biosystems 29(1):1–23
Lin EC, Cantiello HF (1993) A novel method to study the electrodynamic behavior of actin filaments. Evidence for cable-like properties of actin. Biophysical Journal, 65 (December 1992)
Lungarella M, Sporns M (2006) Mapping information flow in sensorimotor networks. PLoS Comput Biol 2(10):e144
Maniotis A, Chen C, Ingber D (1997) Demonstration of mechanical connections between integrins, cytoskeletal filaments and nucleoplasm that stabilise nuclear structure. PNAS 94(3):849–854
Mayne R, Adamatzky A (2015) On the computing potential of intracellular vesicles. PLoS One 10(10):e0139617
Mayne R, Adamatzky A, Jones J (2015a) On the role of the plasmodial cytoskeleton in facilitating intelligent behaviour in slime mould Physarum polycephalum. Commun Integr Biol 8(4):e1059007
Mayne R, Tsompanas M-A, Sirakoulis GC, Adamatzky A (2015b) Towards a slime mould-FPGA interface. Biomed Eng Lett 5(1):51–57
Priel A, Tuszynski J, Woolf N (2010) Neural cytoskeleton capabilities for learning and memory. J Biol Phys 36(1):3–21
Schmidt A, Hall M (1998) Signalling to the actin cytoskeleton. Ann Rev Cell Dev Biol 14:305–338
Stepney S, Braunstein S, Clark J, Tyrrell A, Adamatzky A, Smith R, Addis T, Johnson C, Timmis J, Welch P, Milner R, Partridge D (2005) Journeys in non-classical computation I: a grand challenge for computing research. Int J Parallel Emerg Distrib Syst 20(1):5–19
Tuszyński J, Portet S, Dixon J, Luxford C, Cantiello H (2004) Ionic wave propagation along actin filaments. Biophys J 86:1890–1903
Acknowledgments
This work was supported by the EU research Project Physarum Chip: GrowingComputers from Slime Mould” (FP7 ICT Ref 316366) The authors extend their thanks to Ben De Lacy Costello for his supervision and technical support with the experimental work undertaken, and Dr. Tareq Assaf for his critical insights.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary material 2 (avi 527 KB)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Mayne, R., Adamatkzy, A. Cellular automata modelling of slime mould actin network signalling. Nat Comput 18, 5–12 (2019). https://doi.org/10.1007/s11047-016-9559-0
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11047-016-9559-0