Abstract
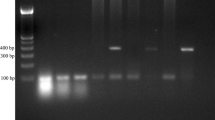
Since C. dubliniensis is similar to C. albicans phenotypically, it can be misidentified as C. albicans. We aimed to investigate the prevalence of C. dubliniensis among isolates previously identified as C. albicans in our stocks and to compare the phenotypic methods and DNA sequencing of D1/D2 region on the ribosomal large subunit (rLSU) gene. A total of 850 isolates included in this study. Phenotypic identification was performed based on germ tube formation, chlamydospore production, colony colors on chromogenic agar, inability of growth at 45 °C and growth on hypertonic Sabouraud dextrose agar. Eighty isolates compatible with C. dubliniensis by at least one phenotypic test were included in the sequence analysis. Nested PCR amplification of D1/D2 region of the rLSU gene was performed after the fungal DNA extraction by Whatman FTA filter paper technology. The sequencing analysis of PCR products carried out by an automated capillary gel electrophoresis device. The rate of C. dubliniensis was 2.35 % (n = 20) among isolates previously described as C. albicans. Consequently, none of the phenotypic tests provided satisfactory performance alone in our study, and molecular methods required special equipment and high cost. Thus, at least two phenotypic methods can be used for identification of C. dubliniensis, and molecular methods can be used for confirmation.
Similar content being viewed by others
References
Miceli MH, Díaz JA, Lee SA. Emerging opportunistic yeast infections. Lancet Infect Dis. 2011;11:142–51.
Sullivan DJ, Westerneng TJ, Haynes KA, Bennett DE, Coleman DC. Candida dubliniensis sp. nov.: phenotypic and molecular characterization of a novel species associated with oral candidosis in HIV-infected individuals. Microbiology. 1995;141:1507–21.
Yu N, Kim HR, Lee MK. The first Korean case of candidemia due to Candida dubliniensis. Ann Lab Med. 2012;32:225–8.
Mubareka S, Vinh DC, Sanche SE. Candida dubliniensis bloodstream infection: a fatal case in a lung transplant recipient. Transpl Infect Dis. 2005;7:146–9.
Horn DL, Neofytos D, Anaissie EJ, et al. Epidemiology and outcomes of candidemia in 2019 patients: data from the prospective antifungal therapy alliance registry. Clin Infect Dis. 2009;48:1695–703.
Loreto ES, Scheid LA, Nogueira CW, Zeni G, Santurio JM, Alves SH. Candida dubliniensis: epidemiology and phenotypic methods for identification. Mycopathologia. 2010;169:431–43.
Alves SH, Milan EP, de Laet Sant’Ana P, Oliveira LO, Santurio JM, Colombo AL. Hypertonic sabouraud broth as a simple and powerful test for Candida dubliniensis screening. Diagn Microbiol Infect Dis. 2002;43:85–6.
Bosco-Borgeat ME, Taverna CG, Cordoba S, et al. Prevalence of Candida dubliniensis fungemia in Argentina: identification by a novel multiplex PCR and comparison of different phenotypic methods. Mycopathologia. 2011;172:407–14.
Ellepola AN, Hurst SF, Elie CM, Morrison CJ. Rapid and unequivocal differentiation of Candida dubliniensis from other Candida species using species-specific DNA probes: comparison with phenotypic identification methods. Oral Microbiol Immunol. 2003;18:379–88.
Ribeiro PM, Querido SM, Back-Brito GN, Mota AJ, Koga-Ito CY, Jorge AO. Research on Candida dubliniensis in a Brazilian yeast collection obtained from cardiac transplant, tuberculosis, and HIV-positive patients, and evaluation of phenotypic tests using agar screening methods. Diagn Microbiol Infect Dis. 2011;71:81–6.
Ahmad S, Khan Z, Asadzadeh M, Theyyathel A, Chandy R. Performance comparison of phenotypic and molecular methods for detection and differentiation of Candida albicans and Candida dubliniensis. BMC Infect Dis. 2012;12:230.
Al-Sweih N, Ahmad S, Khan ZU, Khan S, Chandy R. Prevalence of Candida dubliniensis among germ tube-positive Candida isolates in a maternity hospital in Kuwait. Mycoses. 2005;48:347–51.
Gales AC, Pfaller MA, Houston AK, et al. Identification of Candida dubliniensis based on temperature and utilization of xylose and alpha-methyl-D-glucoside as determined with the API 20C AUX and Vitek YBC systems. J Clin Microbiol. 1999;37:3804–8.
Akgül O, Çerikçioğlu N. Hypertonic sabouraud dextrose agar as a substrate for differentiation of Candida dubliniensis. Mycopathologia. 2009;167:357–9.
Linton CJ, Borman AM, Cheung G, et al. Molecular identification of unusual pathogenic yeast isolates by large ribosomal subunit gene sequencing: 2 years of experience at the United Kingdom mycology reference laboratory. J Clin Microbiol. 2007;45:1152–8.
Borman AM, Linton CJ, Miles SJ, Campbell CK, Johnson EM. Ultra-rapid preparation of total genomic DNA from isolates of yeast and mould using Whatman FTA filter paper technology—a reusable DNA archiving system. Med Mycol. 2007;44:389–98.
Moran GP, Sullivan DJ, Henman MC, et al. Antifungal drug susceptibilities of oral Candida dubliniensis isolates from human immunodeficiency virus (HIV)-infected and non-HIV-infected subjects and generation of stable fluconazole-resistant derivatives in vitro. Antimicrob Agents Chemother. 1997;41:617–23.
Mokaddas E, Khan ZU, Ahmad S. Prevalence of Candida dubliniensis among cancer patients in Kuwait: a 5-year retrospective study. Mycoses. 2011;54:e29–34.
Jabra-Rizk MA, Falkler WA Jr, Merz WG, Baqui AA, Kelley JI, Meiller TF. Retrospective identification and characterization of Candida dubliniensis isolates among Candida albicans clinical laboratory isolates from human immunodeficiency virus (HIV)-infected and non-HIV-infected individuals. J Clin Microbiol. 2000;38:2423–6.
Kim TH, Park BR, Kim HR, Lee MK. Candida dubliniensis screening using the germ tube test in clinical yeast isolates and prevalence of C. dubliniensis in Korea. J Clin Lab Anal. 2010;24:145–8.
Binolfi A, Biasoli MS, Luque AG, Tosello ME, Magaró HM. High prevalence of oral colonization by Candida dubliniensis in HIV-positive patients in Argentina. Med Mycol. 2005;43:431–7.
Odds FC, Van Nuffel L, Dams G. Prevalence of Candida dubliniensis isolates in a yeast stock collection. J Clin Microbiol. 1998;36:2869–73.
Erköse G, Erturan Z. Oral Candida colonization of human immunodeficiency virus infected subjects in Turkey and its relation with viral load and CD4+ T-lymphocyte count. Mycoses. 2007;50:485–90.
Jewtuchowicz VM, Mujica MT, Brusca MI, et al. Phenotypic and genotypic identification of Candida dubliniensis from subgingival sites in immunocompetent subjects in Argentina. Oral Microbiol Immunol. 2008;23:505–9.
Boyanton BL Jr, Luna RA, Fasciano LR, Menne KG, Versalovic J. DNA pyrosequencing-based identification of pathogenic Candida species by using the internal transcribed spacer 2 region. Arch Pathol Lab Med. 2008;132:667–74.
Neppelenbroek KH, Campanha NH, Spolidorio DM, Spolidorio LC, Seó RS, Pavarina AC. Molecular fingerprinting methods for the discrimination between C. albicans and C. dubliniensis. Oral Dis. 2006;12:242–53.
Acknowledgments
This work was supported by a Grant of Turkey Scientific and Technological Research Foundation-TUBITAK (No. 109S129).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kiraz, N., Oz, Y., Aslan, H. et al. The Usefulness of DNA Sequencing After Extraction by Whatman FTA Filter Matrix Technology and Phenotypic Tests for Differentiation of Candida albicans and Candida dubliniensis . Mycopathologia 177, 81–86 (2014). https://doi.org/10.1007/s11046-014-9728-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11046-014-9728-6