Abstract
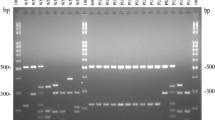
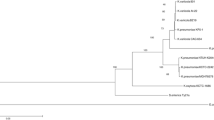
Thirty-seven clinical isolates of C. neoformans were recovered from AIDS patients and all were serotype A according to standard typing tests. They were further analyzed using RAPD, PCR fingerprinting, and PFGE along with 2 additional reference isolates ATCC 34871 (serotype A) and RV 45981 (serotype D). Using 2 different RAPD primers, all of the clinical isolates and the reference serotype A (ATCC 34871) gave similar RAPD patterns while serotype D (RV 45981) gave distinctive pattern. Corresponding result was also obtained upon PCR by using a primer for microsatellite (GACA)4. However, using a primer specific to minisatellite M13 + 1, all PCR fingerprinting gave similar gel patterns (M1) for 35/37 of the clinical isolates and the reference serotype A while two clinical isolates generated different patterns called M2 and M3. The reference serotype D gave distinctive pattern called M4. PFGE gave 17 different karyotypes that could be categorized into 4 groups named EKA (1–6), EKB (1–5), EKC (1– 5) and EKD (1). The reference serotype A fell into group EKA as EKA6 while the reference serotype D fell into group EKC as EKC5. Among the clinical isolates, EKA group (20/37 isolates) and type EKA1 (16/20) dominated with only one isolate each for types EKA2 to EKA5. The next most prevalent was group EKB (12/37 isolates) which dominately fell in type EKB1 (8/12) and only one isolate each for types EKB2 to EKB5. Group EKC (4/37 isolates) and group EKD (1/37) had only one isolate for each type (EKC1 to EKC 4 and EKD1). The 2 predominant karyotypes (EKA1, 16/37 and EKB1, 8/37) may represent two originally common clones of C. neoformans expose among the patients. The high discriminatory power of PFGE infers the benefit of subtyping which lead to better understanding on the epidemiology and pathogenic potential of C. neoformans subtypes. Moreover, PCR fingerprinting and RAPD infer the feasibility of detail analysis between serotypes A and D for unencapsulated C. neoformans.
Similar content being viewed by others
References
RD. Diamond (1991) ArticleTitleThe growing problem of mycoses in patients with the human immunodeficiency virus Rev Infect Dis 13 480–486
S Sukroongreung C Nilakul O Ruangsomboon W Chuakul B. Eampokalap (1996) ArticleTitleSerotypes of Cryptococcus neoformans isolated from patients prior to and during the AIDS era in Thailand Mycopathologia 135 75–78
BP Currie LF Freundlich A. Casadevall (1994) ArticleTitleRestriction fragment length polymorphism analysis of Cryptococcus neoformans isolates from environmental (pigeon excreta) and clinical sources in New York City J Clin Microbiol 32 1188–1192
F Dromer A Varma O Ronin S Mathoulin B. Dupont (1994) ArticleTitleMolecular typing of Cryptococcus neoformans serotype D clinical isolates J Clin Microbiol 32 2364–2371
SCA Chen AG Brownlee TC Sorrell P Ruma DH Ellis T Pfeiffer BR Speed G. Nimmo (1996) ArticleTitleIdentification by random amplification of polymorphic DNA of a common molecular type of Cryptococcus neoformans var. neoformans in patients with AIDS or other immunosuppressive conditions J Infect Dis 173 754–758
W Meyer K Marszewska M Amirmostofian RP Igreja C Hardtke K Methling MA Viviani A Chindamporn S Sukroongreung MA John DH Ellis TC. Sorrell (1999) ArticleTitleMolecular typing of global isolates of Cryptococcus neoformans var. neoformans by polymerase chain reaction fingerprinting and randomly amplified polymorphic DNA-a pilot study to standardize techniques on which to base a detailed epidemiological survey Electrophoresis 20 1790–1799
TC Sorrell SCA Chen P Ruma W Meyer TJ Pfeiffer DH Ellis AG. Brownlee (1996) ArticleTitleConcordance of clinical and environmental isolates of Cryptococcus neoformans var. gattii by random amplification of polymorphic DNA analysis and PCR fingerprinting J Clin Microbiol 34 1253–1260
W Meyer TG Mitchell EZ Freedman R. Vilgalys (1993) ArticleTitleHybridization probes for conventional DNA fingerprinting used as single primers in the polymerase chain reaction to distinguish strains of Cryptococcus neoformans J Clin Microbiol 31 2274–2280
W Meyer TG. Mitchell (1995) ArticleTitlePolymerase chain reaction fingerprinting in fungi using single primers specific to minisatellites and simple repetitive DNA sequence: strain variation in Cryptococcus neoformans Electrophoresis 16 1648–1656
A Varma KJ. Kwon-Chung (1992) ArticleTitleDNA probe for typing of Cryptococcus neoformans J Clin Microbiol 30 2960–2967
A Varma D Swinne F Staib JE Bennett KJ. Kwon-Chung (1995) ArticleTitleDiversity of DNA fingerprints in Cryptococcus neoformans J Clin Microbiol 33 1807–1814
JR Perfect N Ketabchi GM Cox CW Ingram CL. Beiser (1993) ArticleTitleKaryotping of Cryptococcus neoformans as an epidemiological tool J Clin Microbiol 31 3305–3309
ME Klepser MA. Pfaller (1998) ArticleTitleVariation in electrophoretic karyotype and antifungal susceptibility of clinical isolates of Cryptococcus neoformans at a university-affiliated teaching hospital from 1987 to 1994 J Clin Microbiol 36 3653–3656
BL Wickes TD Moore KJ. Kwon-Chung (1994) ArticleTitleComparison of the electrophoretic karyotypes and chromosomal location of ten genes in the two varieties of Cryptococcus neoformans Microbiology 140 IssueIDPt 3 543–550
ME Brandt LC Hutwagner RJ Kuykendall RW Pinner (1995) ArticleTitleThe Cryptococcal Disease Active Surveillance Group. Comparison of multilocus enzyme electrophoresis and random amplified polymorphic DNA analysis for molecular subtyping of Crpytococcus neoformans J Clin Microbiol 33 1890–1895
I Polacheck G Lebens JB. Hicks (1992) ArticleTitleDevelopment of DNA probes for early diagnosis and epidemiological study of cryptococcosis in AIDS patients J Clin Microbiol 30 925–930
S Sukroongreung S Lim S Tantimavanich B Eampokalap D Carter C Nilakul S Kulkeratiyut S. Tansuphaswadikul (2001) ArticleTitlePhenotypic switching and genetic diversity of Cryptococcus neoformans J Clin Microbiol 39 2060–2064
KJ Kwon-Chung I Polacheck JE. Bennett (1982) ArticleTitleImproved diagnostic medium for separation of Cryptococcus neoformans var. neoformans (serotypes A and D) and Cryptococcus neoformans var. gattii (serotypes B and C) J Clin Microbiol 15 535–537
JAH Burik Particlevan RW Schreckhise TC White RA Bowden D. Myerson (1998) ArticleTitleComparison of six extraction techniques for isolation of DNA from filamentous fungi Med Mycol 36 299–303
F Barchiesi RJ Hollis SA Messer G Scalise MG Rinaldi MA. Pfaller (1995) ArticleTitleElectrophoretic karyotype and in-vitro antifungal susceptibility of Cryptococcus neoformans isolates from AIDS patients Diagn Microbiol Infect Dis 23 99–103
T Boekhout A Belkum Particlevan ACAP Leenders HA Verbrugh P Mukamurangwa D Swinne WA. Scheffers (1997) ArticleTitleMolecular typing of Cryptococcus neoformans: taxonomic and epidemiological aspects Int J Syst Bacteriol 47 432–442
H Hotzel P Kielstein R Blaschke-Hellmessen J Wendisch W. Bar (1998) ArticleTitlePhenotypic and genotypic differentiation of several human and avian isolates of Cryptococcus neoformans Mycoses 41 389–396
MA Viviani H Wen A Roverselli R Calderelli-Stefano M Cogliati P Ferrante AM. Tortorano (1997) ArticleTitleIdentification by polymerase chain reaction fingerprinting of Cryptococcus neoformans serotype AD J Med Vet Mycol 35 355–360
A Varma KJ. Kwon-Chung (1989) ArticleTitleRestriction fragment polymorphism in mitochondrial DNA of Cryptococcus neoformans J Gen Microbiol 135 3353–3362
KJ Kwon-Chung BL Wickes L Stockman GD Roberts D Ellis DH. Howard (1992) ArticleTitleVirulence, serotype, and molecular characteristics of environmental strains of Cryptococcus neoformans var. gattii Infect Immun 60 1869–1874
BC Fries FC Chen BP Currie A. Casadevall (1996) ArticleTitleKaryotpe instability in Cryptococcus neoformans infection J Clin Microbiol 34 1531–1534
JB Taylor DM Geiser A Burt V. Koufopanou (1999) ArticleTitleThe evolutionary biology and population genetics underlying fungal strain typing Clin Microbiol Rev 12 126–146
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ngamwongsatit, P., Sukroongreung, S., Nilakul, C. et al. Electrophoretic karyotypes of C. neoformans serotype A recovered from Thai Patients with AIDS. Mycopathologia 159, 189–197 (2005). https://doi.org/10.1007/s11046-004-6671-y
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/s11046-004-6671-y