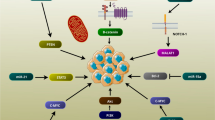
Abstract
Cancer widely affects the world’s health population and ranks second leading cause of death globally. Because of poor prognosis of various types of cancer such as sarcoma, lymphoma, adenomas etc., their high recurrence and metastasis rate and low early diagnosis rate have become concern lately. Role of autophagy in cancer progression is being studied since long. Autophagy is cell’s self-degradative mechanism towards stress and has role in degradation of the cytoplasmic macromolecules which has potential to damage other cytosolic molecules. Autophagy can promote as well as inhibit tumorigenesis depending upon the associated protein combinations in cancer cells. Recent studies have shown that non-coding RNAs (ncRNAs) do not code for protein but play essential role in modulation of gene expression. At transcriptional level, different ncRNAs like lncRNAs, miRNAs and circRNAs directly or indirectly affect different stages of autophagy like autophagy-dependent and non-apoptotic cell death in cancer cells. This review focuses on the involvement of ncRNAs in autophagy and the modulation of several cancer signal transduction pathways in cancers such as lung, breast, prostate, pancreatic, thyroid, and kidney cancer.


Similar content being viewed by others
Abbreviations
- Non-coding RNA:
-
NcRNA
- lncRNA:
-
Long non coding RNA
- miRNA:
-
MicroRNA
- circRNA:
-
Circular RNA
- ATG:
-
Autophagy related gene
- ATG16L1:
-
Autophagy related 16 like 1 protein
- mTOR:
-
Mammalian target of rapamycin
- ULK1:
-
Unc-51 like autophagy activating kinase 1
- FIP200:
-
Focal adhesion kinase family-interacting protein of 200 kDa
- PIP3 kinase III:
-
Phosphatidylinositol 3 kinase III
- LC3II:
-
Microtubule-associated protein 1A/1B-light chain 3
- piRNA:
-
Piwi interacting RNA
- siRNA:
-
Small interfering RNA
- snoRNA:
-
Small nucleolar RNA
- ceRNA:
-
Competing endogenous RNA
- TKI:
-
Tyrosine kinase inhibitor
- NSCLC:
-
Non-small cell lung cancer
- TIGAR:
-
TP53-inducible glycolysis and apoptosis regulator
- ROS:
-
Reactive oxygen species
- DDP:
-
Dipeptidyl peptidase
- EPG2:
-
Ectopic P granules protein 2
- MALAT1:
-
Metastasis-Associated Lung Adenocarcinoma Transcript 1
- HuR:
-
Human antigen R
- PI3K:
-
Phosphoinositide 3-kinase
- GAS5:
-
Growth arrest specific 5 gene
- BLACAT 1:
-
Bladder cancer associated transcript 1
- MRP1:
-
Multidrug resistance associated protein 1
- TSLNC8:
-
Tumor-suppressive lncRNA on chromosome 8p12
- STAT3:
-
Signal transducer and activator of transcription 3
- HIF-1α:
-
Hypoxia inducible factor 1α
- PIK3CB:
-
Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit beta
- SQSTM1:
-
Sequestosome 1
- PRKAA/AMPK:
-
Protein kinase AMP activated catalytic subunit alpha
- TOR:
-
Target of rapamycin
- Wnt:
-
Wingless-related integration site
- BRCA1:
-
Breast cancer gene 1
- PTEN:
-
Phosphatase and Tensin Homolog deleted on chromosome 10
- ELK1:
-
ETS like-1 protien
- ELK3:
-
ETS like-2 protien
- SMAD4:
-
SMAD family member 4
- TGFβ:
-
Transforming growth factor beta
- MAPK:
-
Mitogen activated protein kinase
- XIAP:
-
X-linked inhibitor of apoptosis protein
- HMGB1:
-
High mobility group box 1
- PAX6:
-
Paired box protein 6
- MMPs:
-
Matrix metalloproteinases
- JNK:
-
C-Jun N-terminal kinase 1
- MAPT-AS1:
-
MAPT Antisense RNA 1
- ERK1/2:
-
Extracellular signal regulated kinase 1/2
- LAMP1:
-
Lysosomal-associated membrane protein 1
- RELN:
-
Reclin
- Nrf2:
-
Nuclear factor erythroid related factor 2
- SMC1A:
-
Structural maintenance of chromosome protein 1A
- TRIP13:
-
Thyroid receptor-interacting protein 13
- GAPDH:
-
Glyceraldehyde-3-phosphate dehydrogenase
- MAP1LC3B2:
-
Microtubule associated protein 1 light chain 3 beta 2
- H2AFY:
-
H2A histone family, member Y
- HnRNP-1:
-
Heterogeneous nuclear ribonucleoprotein A1
- EGR1:
-
Early growth response 1 gene
- KIF18A:
-
Kinesin family member 18A
- TUG1:
-
Taurine up-regulated 1
- LKB1:
-
Liver kinase B1
- AMPK:
-
AMP activated protein kinase
- TSPAN1:
-
Tetraspanin 1
- FAM83A:
-
Family member with sequence similarity 83A
- eIF5A2:
-
Eukaryotic translation initiation factor 5A2
- AGR2:
-
Anterior gradient protein 2 homolog
- STK4:
-
Serine/threonine-protein kinase 4
- FOXA1:
-
Forkhead box protein A1
- CHD6:
-
Chromodomain helicase DNA binding protein 6
- GAS8-AS1:
-
GAS8-antisense RNA 1
- LRRK2:
-
Leucine rich repeat kinase 2
- SLC7A11:
-
Solute carrier family 7 member 11
- FOXP3:
-
Foxhead box P3
- FOXE1:
-
Foxhead box E1
- ITGA3:
-
Integrin subunit alpha 3
- ITPR1:
-
Inositol 1,4,5-trisphosphate receptor type 1
- NOX4:
-
NADPH oxidase 4
- CCNB1:
-
Cyclin B1
References
Siegel RL, Miller KD, Fuchs HE, Jemal A (2021) Cancer statistics, 2021. Cancer J Clin 71(1):7–33. https://doi.org/10.3322/caac.21654
Cho YY, Kim DJ, Lee HS, Jeong CH, Cho EJ, Kim MO et al (2013) Autophagy and cellular senescence mediated by Sox2 suppress malignancy of cancer cells. PLoS ONE 8(2):e57172. https://doi.org/10.1371/journal.pone.0057172
Dragowska WH, Weppler SA, Wang JC, Wong LY, Kapanen AI, Rawji JS et al (2013) Induction of autophagy is an early response to gefitinib and a potential therapeutic target in breast cancer. PLoS ONE 8(10):e76503. https://doi.org/10.1371/journal.pone.0076503
Rezaei S, Mahjoubin-Tehran M, Aghaee-Bakhtiari SH, Jalili A, Movahedpour A, Khan H et al (2020) Autophagy-related MicroRNAs in chronic lung diseases and lung cancer. Crit Rev Oncol Hematol. https://doi.org/10.1016/j.critrevonc.2020.103063
Talebian S, Daghagh H, Yousefi B, Ȍzkul Y, Ilkhani K, Seif F, Alivand MR (2020) The role of epigenetics and non-coding RNAs in autophagy: a new perspective for thorough understanding. Mech Ageing Dev. https://doi.org/10.1016/j.mad.2020.111309
Qin Y, Sun W, Zhang H, Zhang P, Wang Z, Dong W, He L, Zhang T, Shao L, Zhang W, Wu C (2018) LncRNA GAS8-AS1 inhibits cell proliferation through ATG5-mediated autophagy in papillary thyroid cancer. Endocrine 59(3):555–564. https://doi.org/10.1007/s12020-017-1520-1
Qin Y, Sun W, Wang Z, Dong W, He L, Zhang T, Shao L, Zhang H (2020) ATF2-induced lncRNA GAS8-AS1 promotes autophagy of thyroid cancer cells by targeting the miR-187-3p/ATG5 and miR-1343-3p/ATG7 axes. Mol Ther Nucl Acids 22:584–600. https://doi.org/10.1016/j.omtn.2020.09.022
Zhang X, He Z, Xiang L, Li L, Zhang H, Lin F, Cao H (2019) Codelivery of GRP78 siRNA and docetaxel via RGD-PEG-DSPE/DOPA/CaP nanoparticles for the treatment of castration-resistant prostate cancer. Drug Des Dev Ther 13:1357. https://doi.org/10.2147/DDDT.S198400
Yang Y, Liang C (2015) MicroRNAs: an emerging player in autophagy. ScienceOpen Res. https://doi.org/10.14293/S2199-1006.1.SOR-LIFE.A181CU.v1
Jiang PC, Bu SR (2019) Clinical value of circular RNAs and autophagy-related miRNAs in the diagnosis and treatment of pancreatic cancer. Hepatobiliary Pancreat Dis Int 18(6):511–516. https://doi.org/10.1016/j.hbpd.2019.09.009
Islam Khan MZ, Tam SY, Law HKW (2019) Autophagy-modulating long non-coding RNAs (LncRNAs) and their molecular events in cancer. Front Genet 9:750. https://doi.org/10.3389/fgene.2018.00750
Doldi V, El Bezawy R, Zaffaroni N (2021) MicroRNAs as epigenetic determinants of treatment response and potential therapeutic targets in prostate cancer. Cancers 13(10):2380. https://doi.org/10.3390/cancers13102380
Wei J, Ge X, Tang Y, Qian Y, Lu W, Jiang K et al (2020) An autophagy-related long noncoding RNA signature contributes to poor prognosis in colorectal cancer. Journal of oncology. https://doi.org/10.1155/2020/4728947
Liu C, Wang JO, Zhou WY, Chang XY, Zhang MM, Zhang Y, Yang XH (2019) Long non-coding RNA LINC01207 silencing suppresses AGR2 expression to facilitate autophagy and apoptosis of pancreatic cancer cells by sponging miR-143-5p. Mol Cell Endocrinol 493:110424. https://doi.org/10.1016/j.mce.2019.04.004
Nam RK, Benatar T, Amemiya Y, Sherman C, Seth A (2020) Mir-139 regulates autophagy in prostate cancer cells through BECLIN-1 and mTOR signaling proteins. Anticancer Res 40(12):6649–6663. https://doi.org/10.21873/anticanres.14689
Son SW, Song MG, Yun BD, Park JK (2021) Noncoding RNAs associated with therapeutic resistance in pancreatic cancer. Biomedicines 9(3):263. https://doi.org/10.3390/biomedicines9030263
Xu P, Wu Q, Yu J, Rao Y, Kou Z, Fang G et al (2020) A systematic way to infer the regulation relations of miRNAs on target genes and critical miRNAs in cancers. Front Genet 11:278. https://doi.org/10.3389/fgene.2020.00278
Rothschild SI, Gautschi O, Batliner J, Gugger M, Fey MF, Tschan MP (2017) MicroRNA-106a targets autophagy and enhances sensitivity of lung cancer cells to Src inhibitors. Lung Cancer 107:73–83. https://doi.org/10.1016/j.lungcan.2016.06.004
Li S, Zeng X, Ma R, Wang L (2018) MicroRNA-21 promotes the proliferation, migration and invasion of non-small cell lung cancer A549 cells by regulating autophagy activity via AMPK/ULK1 signaling pathway. Exp Ther Med 16(3):2038–2045. https://doi.org/10.3892/etm.2018.6370
Chen S, Li P, Li J, Wang Y, Du Y, Chen X et al (2015) MiR-144 inhibits proliferation and induces apoptosis and autophagy in lung cancer cells by targeting TIGAR. Cell Physiol Biochem 35(3):997–1007. https://doi.org/10.1159/000369755
Zhang W, Dong YZ, Du X, Peng XN, Shen QM (2019) MiRNA-153–3p promotes gefitinib-sensitivity in non-small cell lung cancer by inhibiting ATG5 expression and autophagy. Eur Rev Med Pharmacol Sci 23:2444–2452. https://doi.org/10.26355/eurrev_201903_17391
Hua L, Zhu G, Wei J (2018) MicroRNA-1 overexpression increases chemosensitivity of non-small cell lung cancer cells by inhibiting autophagy related 3-mediated autophagy. Cell Biol Int 42(9):1240–1249. https://doi.org/10.1002/cbin.10995
Li H, Liu J, Cao W, Xiao X, Liang L, Liu-Smith F et al (2019) C-myc/miR-150/EPG5 axis mediated dysfunction of autophagy promotes development of non-small cell lung cancer. Theranostics 9(18):5134. https://doi.org/10.7150/thno.34887
Li S, Mei Z, Hu HB, Zhang X (2018) The lncRNA MALAT1 contributes to non-small cell lung cancer development via modulating miR-124/STAT3 axis. J Cell Physiol 233(9):6679–6688. https://doi.org/10.1002/jcp.26325
Li C, Zhao W, Pan X, Li X, Yan F, Liu S et al (2020) LncRNA KTN1-AS1 promotes the progression of non-small cell lung cancer via sponging of miR-130a-5p and activation of PDPK1. Oncogene 39(39):6157–6171. https://doi.org/10.1038/s41388-020-01427-4
Yu X, Ye X, Lin H, Feng N, Gao S, Zhang X, Wang Y, Yu H, Deng X, Qian B (2018) Knockdown of long non-coding RNA LCPAT1 inhibits autophagy in lung cancer. Cancer Biol Med 15(3):228–237. https://doi.org/10.20892/j.issn.2095-3941.2017.0150
Deng X, Feng N, Zheng M, Ye X, Lin H, Yu X et al (2017) PM2 5 exposure-induced autophagy is mediated by lncRNA loc146880 which also promotes the migration and invasion of lung cancer cells. Biochem Biophys Acta 1861(2):112–125. https://doi.org/10.1016/j.bbagen.2016.11.009
Zhang N, Yang GQ, Shao XM, Wei L (2016) GAS5 modulated autophagy is a mechanism modulating cisplatin sensitivity in NSCLC cells. Eur Rev Med Pharmacol Sci 20(11):2271–2277
Huang FX, Chen HJ, Zheng FX, Gao ZY, Sun PF, Peng Q et al (2019) LncRNA BLACAT1 is involved in chemoresistance of non-small cell lung cancer cells by regulating autophagy. Int J Oncol 54(1):339–347. https://doi.org/10.3892/ijo.2018.4614
Yang Y, Jiang C, Yang Y, Guo L, Huang J, Liu X et al (2018) Silencing of LncRNA-HOTAIR decreases drug resistance of non-small cell lung cancer cells by inactivating autophagy via suppressing the phosphorylation of ULK1. Biochem Biophys Res Commun 497(4):1003–1010. https://doi.org/10.1016/j.bbrc.2018.02.141
Zhang L, Wang Y, Xia S, Yang L, Wu D, Zhou Y, Lu J (2020) Long noncoding RNA PANDAR inhibits the development of lung cancer by regulating autophagy and apoptosis pathways. J Cancer 11(16):4783. https://doi.org/10.7150/jca.45291
Fan H, Li J, Wang J, Hu Z (2019) Long non-coding RNAs (lncRNAs) tumor-suppressive role of lncRNA on chromosome 8p12 (TSLNC8) inhibits tumor metastasis and promotes apoptosis by regulating interleukin 6 (IL-6)/signal transducer and activator of transcription 3 (STAT3)/hypoxia-inducible factor 1-alpha (HIF-1α) signaling pathway in non-small cell lung cancer. Med Sci Monitor 25:7624. https://doi.org/10.12659/MSM.917565
Li H, Huang H, Li S, Mei H, Cao T, Lu Q (2021) Long non-coding RNA ADAMTS9-AS2 inhibits liver cancer cell proliferation, migration and invasion. Exp Ther Med 21(6):1–9. https://doi.org/10.3892/etm.2021.9991
Shackelford DB, Shaw RJ (2009) The LKB1–AMPK pathway: metabolism and growth control in tumour suppression. Nat Rev Cancer 9(8):563–575. https://doi.org/10.1038/nrc2676
Kong R (2020) Circular RNA hsa_circ_0085131 is involved in cisplatin-resistance of non-small-cell lung cancer cells by regulating autophagy. Cell Biol Int 44(9):1945–1956. https://doi.org/10.1002/cbin.11401
Fan L, Li B, Li Z, Sun L (2021) Identification of autophagy related circRNA-miRNA-mRNA-subtypes network with radiotherapy responses and tumor immune microenvironment in non-small cell lung cancer. Front Genet. https://doi.org/10.3389/fgene.2021.730003
Zhang F, Cheng R, Li P, Lu C, Zhang G (2021) Hsa_circ_0010235 functions as an oncogenic drive in non-small cell lung cancer by modulating miR-433-3p/TIPRL axis. Cancer Cell Int 21(1):1–14. https://doi.org/10.1186/s12935-021-01764-8
Rahmani F, Tabrizi AT, Hashemian P, Alijannejad S, Rahdar HA, Ferns GA et al (2020) Role of regulatory miRNAs of the Wnt/β-catenin signaling pathway in tumorigenesis of breast cancer. Gene 754:144892. https://doi.org/10.1016/j.gene.2020.144892
Ren Y, Chen Y, Liang X, Lu Y, Pan W, Yang M (2017) MiRNA-638 promotes autophagy and malignant phenotypes of cancer cells via directly suppressing DACT3. Cancer Lett 390:126–136. https://doi.org/10.1016/j.canlet.2017.01.009
Ahmad A, Zhang W, Wu M, Tan S, Zhu T (2018) Tumor-suppressive miRNA-135a inhibits breast cancer cell proliferation by targeting ELK1 and ELK3 oncogenes. Genes Genom 40(3):243–251. https://doi.org/10.1007/s13258-017-0624-6
Cheng Y, Li Z, Xie J, Wang P, Zhu J, Li Y, Wang Y (2018) MiRNA-224-5p inhibits autophagy in breast cancer cells via targeting Smad4. Biochem Biophys Res Commun 506(4):793–798. https://doi.org/10.1016/j.bbrc.2018.10.150
Kandettu A, Radhakrishnan R, Chakrabarty S, Sriharikrishnaa S, Kabekkodu SP (2020) The emerging role of miRNA clusters in breast cancer progression. Biochem Biophys Acta. https://doi.org/10.1016/j.bbcan.2020.188413
Chen P, He YH, Huang X, Tao SQ, Wang XN, Yan H, Wu ZS (2017) MiR-23a modulates X-linked inhibitor of apoptosis-mediated autophagy in human luminal breast cancer cell lines. Oncotarget 8(46):80709. https://doi.org/10.18632/oncotarget.21080
Liu L, He J, Wei X, Wan G, Lao Y, Xu W et al (2017) MicroRNA-20a-mediated loss of autophagy contributes to breast tumorigenesis by promoting genomic damage and instability. Oncogene 36(42):5874–5884. https://doi.org/10.1038/onc.2017.193
Han M, Hu J, Lu P, Cao H, Yu C, Li X et al (2020) Exosome-transmitted miR-567 reverses trastuzumab resistance by inhibiting ATG5 in breast cancer. Cell Death Dis 11(1):1–15. https://doi.org/10.1038/s41419-020-2250-5
Soni M, Patel Y, Markoutsa E, Jie C, Liu S, Xu P, Chen H (2018) Autophagy, cell viability, and chemoresistance are regulated by miR-489 in breast cancer. Mol Cancer Res 16(9):1348–1360. https://doi.org/10.1158/1541-7786.MCR-17-0634
Shi Y, Gong W, Lu L, Wang Y, Ren J (2019) Upregulation of miR-129-5p increases the sensitivity to Taxol through inhibiting HMGB1-mediated cell autophagy in breast cancer MCF-7 cells. Braz J Med Biol Res. https://doi.org/10.1590/1414-431X20198657
Zhang XH, Li BF, Ding J, Shi L, Ren HM, Liu K et al (2020) LncRNA DANCR-miR-758-3p-PAX6 molecular network regulates apoptosis and autophagy of breast cancer cells. Cancer Manage Res 12:4073. https://doi.org/10.2147/CMAR.S254069
Yu Y, Lv F, Liang D, Yang Q, Zhang B, Lin H et al (2017) HOTAIR may regulate proliferation, apoptosis, migration and invasion of MCF-7 cells through regulating the P53/Akt/JNK signaling pathway. Biomed Pharmacother 90:555–561. https://doi.org/10.1016/j.biopha.2017.03.054
Li G, Qian L, Tang X, Chen Y, Zhao Z, Zhang C (2020) Long non-coding RNA growth arrest-specific 5 (GAS5) acts as a tumor suppressor by promoting autophagy in breast cancer. Mol Med Rep 22(3):2460–2468. https://doi.org/10.3892/mmr.2020.11334
Wang D, Li J, Cai F, Xu Z, Li L, Zhu H et al (2019) Overexpression of MAPT-AS1 is associated with better patient survival in breast cancer. Biochem Cell Biol 97(2):158–164. https://doi.org/10.1139/bcb-2018-0039
Liang G, Ling Y, Mehrpour M, Saw PE, Liu Z, Tan W et al (2020) Autophagy-associated circRNA circCDYL augments autophagy and promotes breast cancer progression. Mol Cancer 19(1):1–16. https://doi.org/10.1186/s12943-020-01152-2
Du WW, Yang W, Li X, Awan FM, Yang Z, Fang L et al (2018) A circular RNA circ-DNMT1 enhances breast cancer progression by activating autophagy. Oncogene 37(44):5829–5842. https://doi.org/10.1038/s41388-018-0369-y
Yu X, Li R, Shi W, Jiang T, Wang Y, Li C et al (2016) Silencing of MicroRNA-21 confers the sensitivity to tamoxifen and fulvestrant by enhancing autophagic cell death through inhibition of the PI3K-AKT mTOR pathway in breast cancer cells. Biomed Pharmacother 77:37–44. https://doi.org/10.1016/j.biopha.2015.11.005
Liu LM, Shen WF, Zhu ZH et al (2018) Combined inhibition of EGFR and c-ABL suppresses the growth of fulvestrant resistant breast cancer cells through miR-375-autophagy axis. Biochem Biophys Res Commun 498(3):559–565. https://doi.org/10.1016/j.bbrc.2018.03.019
Zhang FF, Wang BB, Long HL et al (2016) Decreased miR- 124–3p expression prompted breast cancer cell progression mainly by targeting BECLIN-1. Clin Lab 62(6):1139–1145. https://doi.org/10.7754/clin.lab.2015.151111
Pradhan AK, Talukdar S, Bhoopathi P, Shen X-N, Emdad L, Das SK et al (2017) mda-7/IL-24 mediates cancer cell-specific death via regulation of miR- 221 and the BECLIN-1 axis. Cancer Res 77:949–959
Cheng Y, Li Z, Xie J, Wang P, Zhu J, Li Y et al (2018) MiRNA-224-5p inhibits autophagy in breast cancer cells via targeting smad4. Biochem Biophys Res Commun 506:793–798
Chong ZX, Yeap SK, Ho WY (2021) Regulation of autophagy by microRNAs in human breast cancer. J Biomed Sci 28(1):1–18
Lu D, Luo P, Wang Q, Ye Y, Wang B (2017) lncRNA PVT1 in cancer: a review and meta-analysis. Int J Clin Chem 474:1–7. https://doi.org/10.1016/j.cca.2017.08.038
Liao W, Zhang Y (2020) MicroRNA-381 facilitates autophagy and apoptosis in prostate cancer cells via inhibiting the RELN-mediated PI3K/AKT/mTOR signaling pathway. Life Sci 254:117672. https://doi.org/10.1016/j.lfs.2020.117672
Clotaire DZJ, Zhang B, Wei N, Gao R, Zhao F, Wang Y et al (2016) MiR-26b inhibits autophagy by targeting ULK2 in prostate cancer cells. Biochem Biophys Res Commun 472(1):194–200. https://doi.org/10.1016/j.bbrc.2016.02.093
Liu, P. F., Farooqi, A. A., Peng, S. Y., Yu, T. J., Dahms, H. U., Lee, C. H., et al. (2020, October). Regulatory effects of noncoding RNAs on the interplay of oxidative stress and autophagy in cancer malignancy and therapy. In Seminars in Cancer Biology. Academic Press. https://doi.org/10.1016/j.semcancer.2020.10.009
Xiu D, Liu L, Cheng M, Sun X, Ma X (2020) Knockdown of lncRNA TUG1 enhances radiosensitivity of prostate cancer via the TUG1/miR-139-5p/SMC1A axis. Onco Targets Ther 13:2319. https://doi.org/10.2147/OTT.S236860
Yang G, Yin H, Lin F, Gao S, Zhan K, Tong H et al (2020) Long noncoding RNA TUG1 regulates prostate cancer cell proliferation, invasion and migration via the Nrf2 signaling axis. Pathol-Res Practice 216(4):152851. https://doi.org/10.1016/j.prp.2020.152851
Dong L, Ding H, Li Y, Xue D, Liu Y (2018) LncRNA TINCR is associated with clinical progression and serves as tumor suppressive role in prostate cancer. Cancer Manage Res 10:2799. https://doi.org/10.2147/CMAR.S170526
Beaver LM, Kuintzle R, Buchanan A, Wiley MW, Glasser ST, Wong CP et al (2017) Long noncoding RNAs and sulforaphane: a target for chemoprevention and suppression of prostate cancer. J Nutr Biochem 42:72–83. https://doi.org/10.1016/j.jnutbio.2017.01.001
Yang J, Hao T, Sun J, Wei P, Zhang H (2019) Long noncoding RNA GAS5 modulates α-Solanine-induced radiosensitivity by negatively regulating miR-18a in human prostate cancer cells. Biomed Pharmacother 112:108656. https://doi.org/10.1016/j.biopha.2019.108656
Chen C, Wang K, Wang Q, Wang X (2018) LncRNA HULC mediates radioresistance via autophagy in prostate cancer cells. Braz J Med Biol Res. https://doi.org/10.1590/1414-431X20187080
Lu J, Zhong C, Luo J, Shu F, Lv D, Liu Z et al (2021) HnRNP-L-regulated circCSPP1/miR-520h/EGR1 axis modulates autophagy and promotes progression in prostate cancer. Mol Ther-Nucl Acids 26:927–944. https://doi.org/10.1016/j.omtn.2021.10.006
Zhong C, Wu K, Wang S, Long Z, Yang T, Zhong W et al (2021) Autophagy-related circRNA evaluation reveals hsa_circ_0001747 as a potential favorable prognostic factor for biochemical recurrence in patients with prostate cancer. Cell Death Dis 12(8):1–12. https://doi.org/10.1038/s41419-021-04015-w
Cai F, Li J, Zhang J, Huang S (2020) Knockdown of Circ_CCNB2 sensitizes prostate cancer to radiation through repressing autophagy by the miR-30b-5p/KIF18A axis. Cancer Biother Radiopharm. https://doi.org/10.1089/cbr.2019.3538
Borchardt H, Ewe A, Morawski M, Weirauch U, Aigner A (2021) miR24–3p activity after delivery into pancreatic carcinoma cell lines exerts profound tumor-inhibitory effects through distinct pathways of apoptosis and autophagy induction. Cancer Lett 503:174–184. https://doi.org/10.1016/j.canlet.2021.01.018
Gu DN, Jiang MJ, Mei Z, Dai JJ, Dai CY, Fang C et al (2017) microRNA-7 impairs autophagy-derived pools of glucose to suppress pancreatic cancer progression. Cancer Lett 400:69–78. https://doi.org/10.1016/j.canlet.2017.04.020
Ye ZQ, Zou CL, Chen HB, Jiang MJ, Mei Z, Gu DN (2020) MicroRNA-7 as a potential biomarker for prognosis in pancreatic cancer. Dis Markers. https://doi.org/10.1155/2020/2782101
Huang L, Hu C, Cao H, Wu X, Wang R, Lu H et al (2018) MicroRNA-29c increases the chemosensitivity of pancreatic cancer cells by inhibiting USP22 mediated autophagy. Cell Physiol Biochem 47(2):747–758. https://doi.org/10.1159/000490027
Zhou C, Liang Y, Zhou L, Yan Y, Liu N, Zhang R et al (2021) TSPAN1 promotes autophagy flux and mediates cooperation between WNT-CTNNB1 signaling and autophagy via the MIR454-FAM83A-TSPAN1 axis in pancreatic cancer. Autophagy 17(10):3175–3195. https://doi.org/10.1080/15548627.2020.1826689
Wang ZC, Huang FZ, Xu HB, Sun JC, Wang CF (2019) MicroRNA-137 inhibits autophagy and chemosensitizes pancreatic cancer cells by targeting ATG5. Int J Biochem Cell Biol 111:63–71. https://doi.org/10.1016/j.biocel.2019.01.020
Wu Y, Tang Y, Xie S, Zheng X, Zhang S, Mao J et al (2020) Chimeric peptide supramolecular nanoparticles for plectin-1 targeted miRNA-9 delivery in pancreatic cancer. Theranostics 10(3):1151
Huang F, Chen W, Peng J, Li Y, Zhuang Y, Zhu Z et al (2018) LncRNA PVT1 triggers Cyto-protective autophagy and promotes pancreatic ductal adenocarcinoma development via the miR-20a-5p/ULK1 Axis. Mol Cancer 17(1):1–16. https://doi.org/10.1186/s12943-018-0845-6
Yang T, Shen P, Chen Q, Wu P, Yuan H, Ge W et al (2021) FUS-induced circRHOBTB3 facilitates cell proliferation via miR-600/NACC1 mediated autophagy response in pancreatic ductal adenocarcinoma. J Exp Clin Cancer Res 40(1):1–25. https://doi.org/10.1186/s13046-021-02063-w
Wei DM, Jiang MT, Lin P, Yang H, Dang YW, Yu Q et al (2019) Potential ceRNA networks involved in autophagy suppression of pancreatic cancer caused by chloroquine diphosphate: a study based on differentially-expressed circRNAs, lncRNAs, miRNAs and mRNAs. Int J Oncol 54(2):600–626. https://doi.org/10.3892/ijo.2018.4660
Gou L, Zou H, Li B (2019) Long noncoding RNA MALAT1 knockdown inhibits progression of anaplastic thyroid carcinoma by regulating miR-200a-3p/FOXA1. Cancer Biol Ther 20(11):1355–1365. https://doi.org/10.1080/15384047.2019.1617567
Abraham D, Jackson N, Gundara JS, Zhao J, Gill AJ, Delbridge L et al (2011) MicroRNA profiling of sporadic and hereditary medullary thyroid cancer identifies predictors of nodal metastasis, prognosis, and potential therapeutic targets. Clin Cancer Res 17(14):4772–4781. https://doi.org/10.1158/1078-0432.CCR-11-0242
Gundara JS, Zhao J, Gill AJ, Lee JC, Delbridge L, Robinson BG et al (2015) Noncoding RNA blockade of autophagy is therapeutic in medullary thyroid cancer. Cancer Med 4(2):174–182. https://doi.org/10.1002/cam4.355
Zhang Y, Yang WQ, Zhu H, Qian YY, Zhou L, Ren YJ et al (2014) Regulation of autophagy by miR-30d impacts sensitivity of anaplastic thyroid carcinoma to cisplatin. Biochem Pharmacol 87(4):562–570. https://doi.org/10.1016/j.bcp.2013.12.004
Liu J, Feng L, Zhang H, Zhang J, Zhang Y, Li S et al (2018) Effects of miR-144 on the sensitivity of human anaplastic thyroid carcinoma cells to cisplatin by autophagy regulation. Cancer Biol Ther 19(6):484–496. https://doi.org/10.1080/15384047.2018.1433502
Wang S, Wu J, Ren J, Vlantis AC, Li MY, Liu S, Ng E, Chan A, Luo DC, Liu Z, Guo W, Xue L, Ng SK, van Hasselt CA, Tong M, Chen GG (2018) MicroRNA-125b interacts with Foxp3 to induce autophagy in thyroid cancer. Mol Ther 26(9):2295–2303. https://doi.org/10.1016/j.ymthe.2018.06.015
Liu H, Chen X, Lin T, Chen X, Yan J, Jiang S (2019) MicroRNA-524-5p suppresses the progression of papillary thyroid carcinoma cells via targeting on FOXE1 and ITGA3 in cell autophagy and cycling pathways. J Cell Physiol 234(10):18382–18391. https://doi.org/10.1002/jcp.28472
Yang LX, Wu J, Guo ML, Zhang Y, Ma SG (2019) Suppression of long non-coding RNA TNRC6C-AS1 protects against thyroid carcinoma through DNA demethylation of STK4 via the Hippo signalling pathway. Cell Prolif 52(3):e12564. https://doi.org/10.1111/cpr.12564
Peng X, Ji C, Tan L, Lin S, Zhu Y, Long M, Luo D, Li H (2020) Long non-coding RNA TNRC6C-AS1 promotes methylation of STK4 to inhibit thyroid carcinoma cell apoptosis and autophagy via Hippo signalling pathway. J Cell Mol Med 24(1):304–316. https://doi.org/10.1111/jcmm.14728
Mila G, Manicardi V, Federica T, Elisabetta S, Riccardo B, Gloria M et al (2021) Linc00941 is a novel TGFβ target that primes papillary thyroid cancer metastatic behavior by regulating the expression of Cadherin. Thyroid. https://doi.org/10.1089/thy.2020.0001
Zhao Y, Zhao L, Li J, Zhong L (2019) Silencing of long noncoding RNA RP11-476D10. 1 enhances apoptosis and autophagy while inhibiting proliferation of papillary thyroid carcinoma cells via microRNA-138-5p-dependent inhibition of LRRK2. J Cell Physiol 234(11):20980–20991. https://doi.org/10.1002/jcp.28702
Wang HH, Ma JN, Zhan XR (2021) Circular RNA Circ_0067934 attenuates ferroptosis of thyroid cancer cells by miR-545–3p/SLC7A11 signaling. Front Endocrinol. https://doi.org/10.3389/fendo.2021.670031
Liu F, Zhang J, Qin L, Yang Z, Xiong J, Zhang Y et al (2018) Circular RNA EIF6 (Hsa_circ_0060060) sponges miR-144–3p to promote the cisplatin-resistance of human thyroid carcinoma cells by autophagy regulation. Aging 10(12):3806. https://doi.org/10.18632/aging.101674
Shi YP, Liu GL, Li S, Liu XL (2020) miR-17-5p knockdown inhibits proliferation, autophagy and promotes apoptosis in thyroid cancer via targeting PTEN. Neoplasma 67(2):249–258. https://doi.org/10.4149/neo_2019_190110N29
Gundara JS, Zhao J, Gill AJ, Lee JC, Delbridge L, Robinson BG, McLean C, Serpell J, Sidhu SB (2015) Noncoding RNA blockade of autophagy is therapeutic in medullary thyroid cancer. Cancer Med 4(2):174–182. https://doi.org/10.1002/cam4.355
Zhao Y, Zhao L, Li J, Zhong L (2019) Silencing of long noncoding RNA RP11–476D10.1 enhances apoptosis and autophagy while inhibiting proliferation of papillary thyroid carcinoma cells via microRNA-138–5p-dependent inhibition of LRRK2. J Cell Physiol 234(11):20980–20991. https://doi.org/10.1002/jcp.28702
Liu J, Feng L, Zhang H, Zhang J, Zhang Y, Li S, Qin L, Yang Z, Xiong J (2018) Effects of miR-144 on the sensitivity of human anaplastic thyroid carcinoma cells to cisplatin by autophagy regulation. Cancer Biol Ther 19(6):484–496. https://doi.org/10.1080/15384047.2018.1433502
Wen D, Liu WL, Lu ZW, Cao YM, Ji QH, Wei WJ (2021) SNHG9, a papillary thyroid cancer cell exosome-enriched lncRNA, inhibits cell autophagy and promotes cell apoptosis of normal thyroid epithelial cell Nthy-ori-3 through YBOX3/P21 pathway. Front Oncol 11:647034. https://doi.org/10.3389/fonc.2021.647034
Peng D, Li W, Zhang B, Liu X (2021) Overexpression of lncRNA SLC26A4-AS1 inhibits papillary thyroid carcinoma progression through recruiting ETS1 to promote ITPR1-mediated autophagy. J Cell Mol Med 25(17):8148–8158. https://doi.org/10.1111/jcmm.16545
Chen Y, Zhou J, Wu X, Huang J, Chen W, Liu D, Zhang J, Huang Y, Xue W (2020) miR-30a-3p inhibits renal cancer cell invasion and metastasis through targeting ATG12. Transl Androl Urol 9(2):646–653. https://doi.org/10.21037/tau.2019.12.10
Liu X, Zhong L, Li P, Zhao P (2020) MicroRNA-100 enhances autophagy and suppresses migration and invasion of renal cell carcinoma cells via disruption of NOX4-dependent mTOR pathway. Clin Transl Sci. https://doi.org/10.1111/cts.12798
Zheng B, Zhu H, Gu D, Pan X, Qian L, Xue B, Yang D, Zhou J, Shan Y (2015) MiRNA-30a-mediated autophagy inhibition sensitizes renal cell carcinoma cells to sorafenib. Biochem Biophys Res Commun 459(2):234–239. https://doi.org/10.1016/j.bbrc.2015.02.084
Takai T, Tsujino T, Yoshikawa Y, Inamoto T, Sugito N, Kuranaga Y, Heishima K, Soga T, Hayashi K, Miyata K, Kataoka K, Azuma H, Akao Y (2019) Synthetic miR-143 exhibited an anti-cancer effect via the downregulation of K-RAS networks of renal cell cancer cells in vitro and in vivo. Mol Ther 27(5):1017–1027. https://doi.org/10.1016/j.ymthe.2019.03.004
Mikhaylova O, Stratton Y, Hall D, Kellner E, Ehmer B, Drew AF, Gallo CA, Plas DR, Biesiada J, Meller J, Czyzyk-Krzeska MF (2012) VHL-regulated MiR-204 suppresses tumor growth through inhibition of LC3B-mediated autophagy in renal clear cell carcinoma. Cancer Cell 21(4):532–546. https://doi.org/10.1016/j.ccr.2012.02.019
Yan L, Liu G, Cao H, Zhang H, Shao F (2019) Hsa_circ_0035483 sponges hsa-miR-335 to promote the gemcitabine-resistance of human renal cancer cells by autophagy regulation. Biochem Biophys Res Commun 519(1):172–178. https://doi.org/10.1016/j.bbrc.2019.08.093
Jin Y, Huang R, Xia Y, Huang C, Qiu F, Pu J, He X, Zhao X (2020) Long noncoding RNA KIF9-AS1 regulates transforming growth factor-β and autophagy signaling to enhance renal cell carcinoma chemoresistance via microRNA-497-5p. DNA Cell Biol 39(7):1096–1103. https://doi.org/10.1089/dna.2020.5453
Shao Q, Wang Q, Wang J (2019) LncRNA SCAMP1 regulates ZEB1/JUN and autophagy to promote pediatric renal cell carcinoma under oxidative stress via miR-429. Biomed Pharmacother 120:109460. https://doi.org/10.1016/j.biopha.2019.109460
Li D, Li C, Chen Y, Teng L, Cao Y, Wang W, Pan H, Xu Y, Yang D (2020) LncRNA HOTAIR induces sunitinib resistance in renal cancer by acting as a competing endogenous RNA to regulate autophagy of renal cells. Cancer Cell Int 20:338. https://doi.org/10.1186/s12935-020-01419-0
Su Y, Lu J, Chen X, Liang C, Luo P, Qin C, Zhang J (2019) Long non-coding RNA HOTTIP affects renal cell carcinoma progression by regulating autophagy via the PI3K/Akt/Atg13 signaling pathway. J Cancer Res Clin Oncol 145(3):573–588. https://doi.org/10.1007/s00432-018-2808-0
Gui CP, Cao JZ, Tan L, Huang Y, Tang YM, Li PJ, Chen YH, Lu J, Yao HH, Chen ZH, Pan YH, Ye YL, Qin ZK, Chen W, Wei JH, Luo JH (2021) A panel of eight autophagy-related long non-coding RNAs is a good predictive parameter for clear cell renal cell carcinoma. Genomics 113(2):740–754. https://doi.org/10.1016/j.ygeno.2021.01.016
Sahin Lacin EE, Karakas Y, Yalcin S (2015) Metastatic medullary thyroid cancer: a dramatic response to a systemic chemotherapy (temozolomide and capecitabine) regimen. OncoTargets Ther 8:1039. https://doi.org/10.2147/OTT.S82906
Funding
This study was not funded by any authority.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Barnwal, S.K., Bendale, H. & Banerjee, S. Non-coding RNAs associated with autophagy and their regulatory role in cancer therapeutics. Mol Biol Rep 49, 7025–7037 (2022). https://doi.org/10.1007/s11033-022-07517-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-022-07517-8