Abstract
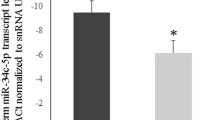
MicroRNAs are small, non-coding, single-strand oligonucleotides which regulate gene expression. There is little evidence in the literature about their role in azoospermia and no studies have investigated their presence in the seminal plasma of men with Klinefelter syndrome. This retrospective study investigated if there were any differences in microRNA expression (miR-509-5p, miR-122-5p, miR-34b-3p, miR-34c-5p) in the seminal plasma of patients with obstructive azoospermia, non-obstructive azoospermia and Klinefelter syndrome. Hormone levels were also investigated to identify any correlations with microRNA expression. We analysed 200 subjects (40 Klinefelter syndrome, 60 non-obstructive azoospermia with a normal karyotype, 60 obstructive azoospermia and 40 who were normozoospermic). All subjects underwent semen examination. Total RNA was obtained from seminal plasma and microRNA expression was analysed by RT-qPCR. There was a significant reduction in the expression of all investigated miRNAs in the seminal plasma of all patient categories in comparison with controls. There was a weak negative correlation between FSH values and miR-509-5p expression in non-obstructive azoospermic patients (r = − 0.391; p = 0.014). We hypothesize that in non-obstructive azoospermia and Klinefelter syndrome patients, the downregulation of microRNAs may be caused by damage to the germ cells and aberrant spermatogenesis. In our opinion the identification of seminal plasma microRNAs deriving almost exclusively from the testes could be essential for the development of specific biomarkers for male infertility. The expression of such microRNAs, in combination with hormone values, could comprise testicular markers of abnormal spermatogenesis and failed mature sperm production.


Similar content being viewed by others
References
World Health Organization (2010) WHO laboratory manual for the examination and processing of human semen, 5th edn. World Health Organization, Geneva, pp 669–676
Ramasamy R, Schlegel PN (2007) microdissection testicular sperm extraction: effect of prior biopsy on success of sperm retrieval. J Urol 177:1447–1449
Wosnitzer M, Goldstein M, Hardy MP (2014) Review of Azoospermia Spermatogenesis 4:e28218
Vogt PH, Edelmann A, Kirsch S, Henegariu O, Hirschmann P et al (1996) Human Y chromosome azoospermia factors (AZF) mapped to different subregions in Yq11. Hum Mol Genet 5:933–943
Tahani N, Nieddu L, Prossomariti G, Spaziani M, Granato S et al (2018) Long-term effect of testosterone replacement therapy on bone in hypogonadal men with Klinefelter Syndrome. Endocrine 61:327–335
Kamischke A, Baumgardt A, Horst J, Nieschlag E (2003) Clinical and diagnostic features of patients with suspected Klinefelter syndrome. J Androl 24:41–48
Aksglaede L, Wikström AM, Rajpert-De Meyts E, Dunkel L, Skakkebaek NE et al (2006) Natural history of seminiferous tubule degeneration in Klinefelter syndrome. Hum Reprod Update 12:39–48
Ambros V (2004) The functions of animal microRNAs. Nature 431:350–355
Abu-Halima M, Backes C, Leidinger P, Keller A, Lubbad AM et al (2014) MicroRNA expression profiles in human testicular tissues of infertile men with different histopathologic patterns. Fertil Steril 101:78–86
Pelloni M, Coltrinari G, Paoli D, Pallotti F, Lombardo F et al (2016) Differential expression of miRNAs in the seminal plasma and serum of testicular cancer patients. Endocrine 57:518–527
Lee YS, Kim HK, Chung S, Kim KS, Dutta A (2005) Depletion of human micro-RNA miR-125b reveals that it is critical for the proliferation of differentiated cells but not for the down-regulation of putative targets during differentiation. J Biol Chem 280:16635–16641
Chen CZ, Li L, Lodish HF, Bartel DP (2004) MicroRNAs modulate hematopoietic lineage differentiation. Science 303:83–86
Ambros V (2003) MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell 113:673–676
Esquela-Kerscher A, Slack FJ (2006) Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer 6:259–269
Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH et al (2010) The microRNA spectrum in 12 body fluids. Clin Chem 56:1733–1741
Wang C, Yang C, Chen X, Yao B, Yang C et al (2011) Altered profile of seminal plasma microRNAs in the molecular diagnosis of male infertility. Clin Chem 57:1722–1731
Kotaja N (2014) MicroRNAs and spermatogenesis. Fertil Steril 101:1552–1562
Hayashi K, de Sousa C, Lopes SM, Kaneda M, Tang F, Hajkova P et al (2008) MicroRNA biogenesis is required for mouse primordial germ cell development and spermatogenesis. PLoS ONE 3:e1738
Papaioannou MD, Nef S (2010) microRNAs in the testis: building up male fertility. J Androl 31:26–33
Barad O, Meiri E, Avniel A, Aharonov R, Barzilai A et al (2004) MicroRNA expression detected by oligonucleotide microarrays: system establishment and expression profiling in human tissues. Genome Res 14:2486–2494
Ro S, Park C, Sanders KM, McCarrey JR, Yan W (2007) Cloning and expression profiling of testis-expressed microRNAs. Dev Biol 311:592–602
Guo X, Su B, Zhou Z, Sha J (2009) Rapid evolution of mammalian X-linked testis microRNAs. BMC Genomics 10:97
Yu Z, Raabe T, Hecht NB (2005) MicroRNA Mirn122a reduces expression of the posttranscriptionally regulated germ cell transition protein 2 (Tnp2) messenger RNA (mRNA) by mRNA cleavage. Biol Reprod 73:427–433
Liu T, Huang Y, Liu J, Zhao Y, Jiang L et al (2013) MicroRNA-122 influences the development of sperm abnormalities from human induced pluripotent stem cells by regulating TNP2 expression. Stem Cells Dev 22:1839–1850
Lin CJF, Gong HY, Tseng HC, Wang WL, Wu JL (2008) miR-122 targets an anti-apoptotic gene, Bcl-w, in human hepatocellular carcinoma cell lines. Biochem Biophys Res Commun 375:315–320
Hermeking H (2009) The miR-34 family in cancer and apoptosis. Cell Death Differ 17:193–199
He L, He X, Lim LP, de Stanchina E, Xuan Z et al (2007) A microRNA component of the p53 tumour suppressor network. Nature 447:1130–1134
Kong YW, Cannell IG, de Moor CH, Hill K, Garside PG et al (2008) The mechanism of micro-RNA-mediated translation repression is determined by the promoter of the target gene. Proc Natl Acad Sci USA 105:8866–8871
Wei JS, Song YK, Durinck S, Chen QR, Cheuk AT et al (2008) The MYCN oncogene is a direct target of miR-34a. Oncogene 27:5204–5213
Christoffersen NR, Shalgi R, Frankel LB, Leucci E, Lees M et al (2010) p53-independent upregulation of miR-34a during oncogene-induced senescence represses MYC. Cell Death Differ 17:236–245
Pulikkan JA, Peramangalam PS, Dengler V, Ho PA, Preudhomme C et al (2010) C/EBPa regulated microRNA-34a targets E2F3 during granulopoiesis and is down-regulated in AML with CEBPA mutations. Blood 116:5638–5649
Rokavec M, Li H, Jiang L, Hm H (2014) The p53/miR-34 axis in development and disease. J Mol Cell Biol 6:214–230
Yan N, Lu Y, Sun H, Tao D, Zhang S et al (2007) A microarray for microRNA profiling in mouse testis tissues. Reproduction 134:73–79
Yan N, Lu Y, Sun H, Qui W, Tao D et al (2009) Microarray profiling of microRNAs expressed in testis tissues of developing primates. J Assist Reprod Genet 26:179–186
Bouhallier F, Allioli N, Lavial F, Chalmel F, Perrard MH et al (2010) Role of miR-34c microRNA in the late steps of spermatogenesis. RNA 16:720–731
Liang X, Zhou D, Wei C, Luo H, Liu J et al (2012) MicroRNA-34c enhances murine male germ cell apoptosis through targeting ATF1. PLoS ONE 7:e33861
Zhang S, Yu M, Liu C, Wang L, Hu Y et al (2012) MiR-34c regulates mouse embryonic stem cells differentiation into male germ-like cells through RARg. Cell biochem and function 30:623–632
Yang Q, Hua J, Wang L, Xu B, Zhang H et al (2013) MicroRNA and piRNA profiles in normal human testis detected by next generation sequencing. PLoS ONE 8:e66809
Liu WM, Pang RT, Chiu PC, Wong BP, Lao K et al (2012) Sperm-borne microRNA-34c is required for the first cleavage division in mouse. Proc Natl Acad Sci U S A 109:490–494
Yu M, Mu H, Niu Z, Chu Z, Zhu H et al (2014) miR-34c enhances muose spermatogonial stem cells differentiation by targeting Nanos2. J Cell Biochem 115:232–242
Song YH, Wang J, Nie G, Chen YJ, Li X et al (2017) MicroRNA-509-5p functions as an anti-oncogene in breast cancer via targeting SOD2. Eur Rev Med Pharmacol Sci 21:3617–3625
Hu L, Wu C, Guo C, Li H, Xiong C (2014) Identification of microRNAs predominately derived from testis and epididymis in human seminal plasma. Clin Biochem 47:967–972
Abu-Halima M, Hammadeh M, Schmitt J, Leidinger P, Keller A et al (2013) Altered microRNA expression profiles of human spermatozoa in patients with different spermatogenic impairments. Fertil Steril 99:1249–1255
Barceló M, Mata A, Bassas L, Larriba S (2018) Exosomal microRNAs in seminal plasma are markers of the origin of azoospermia and can predict the presence of sperm in testicular tissue. Hum Reprod 33:1087–1098
Sui W, Ou M, Chen J, Li H, Lin H et al (2012) microRNA expression profile in peripheral blood mononuclear cells of Klinefelter syndrome. Exp Ther Med 4:825–831
Cimino L, Salemi M, Cannarella R, Condorelli RA, Giurato G et al (2017) Decreased miRNA expression in Klinefelter syndrome. Sci Rep 30:16672
Procópio MS, de Avelar GF, Costa GMJ, Lacerda SMSN, Resende RR et al (2017) MicroRNAs in Sertoli cells: implications for spermatogenesis and fertility. Cell Tissue Res 370:335–346
Nicholls PK, Harrison CA, Walton KL, McLachlan RI, O'Donnell L et al (2011) Hormonal regulation of sertoli cell micro-RNAs at spermiation. Endocrinology 152:1670–1683
Williams M, Cheng YY, Blenkiron C, Reid G (2017) Exploring Mechanisms of MicroRNA Downregulation in Cancer. Microrna 6:2–16
Jun HH, Kwack K, Lee KH, Kim JO, Park HS, Ryu CS, Lee JY, Ko D, Kim J, Kim NK (2019) Association between TP53 genetic polymorphisms and the methylation and expression of miR-34a, 34b/c in colorectal cancer tissues. Oncol Lett. 17(5):4726–4734
Wang Z, Chen Z, Gao Y, Li N, Li B, Tan F, Tan X, Lu N, Sun Y, Sun J et al (2011) DNA hypermethylation of microRNA-34b/c has prognostic value for stage I non-small cell lung cancer. Cancer Biol Ther 11:490–496
Tanaka N, Toyooka S, Soh J, Kubo T, Yamamoto H, Maki Y, Muraoka T, Shien K, Furukawa M, Ueno T et al (2012) Frequent methylation and oncogenic role of microRNA-34b/c in small-cell lung cancer. Lung Cancer 76:32–38
Xie K, Liu J, Chen J, Dong J, Ma H, Liu Y, Hu Z. Methylation-associated silencing of microRNA-34b in hepatocellular carcinoma cancer. Gene. 2014;543(1):101–107.
Suzuki H, Yamamoto E, Nojima M, Kai M, Yamano HO, Yoshikawa K, Kimura T, Kudo T, Harada E, Sugai T et al (2010) Methylation-associated silencing of microRNA-34b/c in gastric cancer and its involvement in an epigenetic field defect. Carcinogenesis 31:2066–2073
Kozaki K, Imoto I, Mogi S, Omura K, Inazawa J (2008) Exploration of tumor-suppressive microRNAs silenced by DNA hypermethylation in oral cancer. Cancer Res 68:2094–2105
Momeni A, Najafipour R, Hamta A, Jahani S, Moghbelinejad S (2020) Expression and Methylation Pattern of hsa-miR-34 Family in Sperm Samples of Infertile Men. Reprod Sci 27(1):301–308
Li Y, Ren Q, Zhu L, Li Y, Li J, Zhang Y, Zheng G, Han T, Sun S, Feng F (2018) Involvement of methylation of MicroRNA-122, -125b and -106b in regulation of Cyclin G1, CAT-1 and STAT3 target genes in isoniazid-induced liver injury. BMC Pharmacol Toxicol 19(1):11
Feng W, Chakraborty A (2017) Fragility extraordinaire: unsolved mysteries of chromosome fragile sites. Adv Exp Med Biol 1042:489–526
Marquardt S, Richter C, Pützer BM, Logotheti S (2020) MiRNAs targeting double strand DNA repair pathways lurk in genomically unstable rare fragile sites and determine cancer outcomes. Cancers (Basel). 12(4):876
Zhang R, Peng Y, Wang W, Su B (2007) Rapid evolution of an X-linked microRNA cluster in primates. Genome Res 17:612–617
Li J, Liu Y, Dong D, Zhang Z (2010) Evolution of an X-linked primate-specific micro RNA cluster. Mol Biol Evol 27:671–683
Sun Z, Zhang Y, Zhang R, Qi X, Su B (2013) Functional divergence of the rapidly evolving miR-513 subfamily in primates. BMC Evol Biol 13:255
Dillon LW, Burrow AA, Wang Y-H (2010) DNA instability at chromosomal fragile sites in cancer. Curr Genomics 11:326–337
Acknowledgements
The authors wish to thank Marie-Hélène Hayles for her assistance in the English translation of the manuscript.
Funding
Supported by a Grant from the Italian Ministry of Education and Research (MIUR-PRIN 2015-2015XSNA83-002) and the University of Rome “Sapienza” Faculty of Medicine.
Author information
Authors and Affiliations
Contributions
DP conceived and designed the experiments; FF and MP wrote the article; FF and MP acquired and analysed the data; DP, FP and FL contributed to data interpretation; AFR, GB and AL and FL contributed to manuscript revision. All authors approved the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interest.
Ethical approval
This retrospective study was approved by our University Hospital’s institutional review board (Protocol 835/18).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Finocchi, F., Pelloni, M., Balercia, G. et al. Seminal plasma miRNAs in Klinefelter syndrome and in obstructive and non-obstructive azoospermia. Mol Biol Rep 47, 4373–4382 (2020). https://doi.org/10.1007/s11033-020-05552-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-020-05552-x