Abstract
Fragments of four candidate reference genes of Aphelenchoides besseyi, including actin, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), ubiquitin conjugating-3 enzyme (UBC) and alpha-tubulin (α-tubulin) were cloned from the transcriptome database of A. besseyi. The expression level of these four candidate reference genes and a commonly used reference gene of A. besseyi (18S rRNA) in three experimental conditions, including the four life stages (female, male, juvenile and egg) of two populations and the mixed-stage nematodes of four populations with different origins and hosts were analyzed by RT-qPCR. The expression stability of the five candidate reference genes under the three experimental conditions was analyzed by ΔCt, geNorm, NormFinder and RefFinder respectively. The analysis results of ΔCt, geNorm, NormFinder and RefFinder all indicated that UBC was the gene with the highest average ranking of stability. In conclusion, the expression stability of UBC was optimal under the three experimental conditions, indicating that UBC could be used as a suitable reference gene instead of 18S rRNA in the RT-qPCR analysis for A. besseyi.
Similar content being viewed by others
Introduction
Quantitative real time PCR (qPCR) is a robust nucleic acid quantitative technique developed on the basis of traditional PCR technology, in which the fluorescent group is added into the PCR reaction system to detect the change of the product quantity by detecting fluorescence intensity change [1]. Compared with traditional PCR, it’s quantitative with an enormous dynamic window. It has become one of the most common techniques in gene expression quantification and transcriptome analysis. A reference gene, also known as a housekeeping gene, is a gene that is expressed in all cells and its expression level is little affected by environmental factors and remains consistent among growth stages, organs and tissues (Pfaffl [2]). When using reverse-transcription qPCR (real-time RT-PCR, RT-qPCR) to analyze the relative quantitative expression of target genes, the incorporation of a reference gene is helpful to correct for any deviations in the initial template amount, RNA quality and enzymatic reaction efficiency among different tissues and cells. By using the reference genes to normalize and standardize the expression level of the target gene, errors in the experimental process can be reduced, thus improving the reliability of the results [3, 4]. The ideal reference gene has constant expression under various experimental conditions, such as different life stages or different populations of nematodes. However, recent studies have shown that there is no perfect reference gene, as experimental conditions change, some traditional reference gene’s expression level change as well. Therefore, under a specific set of experimental conditions, the use of an inappropriate reference gene can lead to inaccurate quantitative results [5, 6]. In recent years, some studies have been conducted to identify appropriate reference genes for different kinds of nematodes. Twelve reference genes were selected to determine their expression stability in Caenorhabditis elegans under different experimental conditions. The results showed that cdc-42, pmp-3 and Y45F10D.4 were relatively stable reference genes (Hoogewijs et al. [7]). The expression stability of 11 reference genes was studied between females and males of different genotypes of Haemonchus contortus and gpd, ama and far were identified as good reference genes [8]. Eleven candidate reference genes were chosen from RNA-seq data of Globodera rostochiensis to identify a reliable set of reference genes to study gene expression, three genes including GR, PMP-3 and aaRS were found to be stable in different stages of G.rostochiensis (Sabeh et al. [9]). eIF5a were identified as a suitable reference gene that could be expressed stably in different developmental stages and different populations of Radopholus similis [10]. Such studies confirmed that not all traditional reference genes are suitable for a qPCR relative quantitative analysis under all experimental conditions; the most suitable reference gene may change as the experimental conditions change. At present, there are no other reports on the reference gene screening of plant nematodes except R. similis and G. rostochiensis, so the research on this field remain further study.
Aphelenchoides besseyi, which is widely distributed in most rice-growing areas throughout the world, is a foliar nematode that feeds endoparasitically or ectoparasitically on the above-ground parts of plants and is one of the most damaging plant nematodes to rice [11]. A. besseyi also endangers 35 genera of higher plants, including strawberries, maize and chestnut, and it is considered to be one of the ten most important plant parasitic nematodes [12]. At present, there are few studies on the reference genes of A. besseyi, and only 18S rRNA was used as reference gene in RT-qPCR analysis [13, 14]. However, there are several arguments against the use of 18S rRNA as a suitable reference gene. First, the expression level of 18S rRNA in cells is too high to deduct the baseline value for a target gene with low expression level in RT-qPCR experiments, which might lead to inaccurate results. Second, oligo dt primers are commonly used in reverse transcription of target genes, while random primers must be used in reverse transcription experiments of 18S rRNA due to the absence of polyA, which may cause deviation due to different reverse transcription efficiency. Third, the gene type of the reference genes and target genes should be close; however, 18S rRNA is a kind of ribosomal RNA gene whereas most target genes are mRNA genes. The effect of the partial degradation of RNA on the expression level of 18S rRNA is typically less than that of the target gene, which makes 18S rRNA unsuitable for monitoring the target gene [6, 15,16,17]. Fourth, some nematode species of Aphalenchoides, in particular A. besseyi and the closely related A. fujianensis has been reported as having variation in ribosomal subunit copy number between species and populations [18, 19], suggesting that the expression level of 18S rRNA may vary greatly between different populations. Therefore, it’s necessary to find a better reference gene to replace 18S rRNA for A. besseyi expression studies.
ΔCt [20], geNorm [21], NormFinder [22] and RefFinder [23] are the four main types of software used to analyze the stability of reference genes. ΔCT method calculates the expression quantity (Q) according to the formula Q = 2ΔCt, which will be a basis for the software to rank the expression stability of each reference gene [20]. For geNorm, the default value of the software M = 1.5 is used to calculate average expression stability value (M) of each reference gene. If the M value of the candidate reference gene is less than 1.5, it can be considered as an available reference gene. In addition, geNorm determines the effect of introducing new reference genes by pairwise variation analysis (Vn/Vn + 1 and determines the optimal number of reference genes; geNorm software uses Vn/Vn + 1 = 0.15 as the cut-off value, if Vn/Vn + 1 < 0.15, it means that (n) genes have been stable as a combination with no need for introducing the (n + 1) reference genes. However, the geNorm operation manual states that the default cut-off value is not fixed and can be adjusted slightly according to the biological and experimental conditions [21]. Because the value Vn/Vn + 1 were all greater than 0.15 under one of the three experimental conditions in this study, the cut-off value Vn/Vn + 1 was set to 0.2 and the optimal number of reference genes is selected under the condition of Vn/Vn + 1 < 0.2. Similar to geNorm, NormFinder can calculate the average expression stability value M of each candidate reference gene, the smaller the M value, the higher the expression stability of a reference gene [22]. The online program RefFinder (https://www.heartcure.com.au/reffinder/?type=reference) integrates the evaluation results of multiple reference gene analysis softwares such as ΔCt, geNorm, and NormFinder to give each analysis method a certain weight. A higher ranking indicates a higher stability of a reference gene [23].
In recent years, using transcriptome database to screen reference genes has been proved to be an effective method, which has been applied to yeast [24], fungi [25], animals [26, 9, 27] and plants [28]. In our previous work [29], in order to understand the developmental and reproductive characteristics of A. besseyi at the molecular level and the mechanism of interaction between A. besseyi and host plants, we have generated a transcriptome database (accession number: SRR4002929 and SRR4002930) consisting of two populations of A. besseyi with different pathogenicity. In this study, the four commonly used reference genes actin, GAPDH, UBC and α-tubulin were cloned from this transcriptome database. The expression stability of these four candidate genes and 18S rRNA in four life stages of two populations and mixed-stage of four populations of A. besseyi were studied by four commonly used reference gene screening software including ΔCt, geNorm, NormFinder and RefFinder. The results of this study confirmed that UBC is an ideal reference gene in RT-qPCR experiments of A. besseyi, which laid a solid foundation for accurate quantification of A. besseyi genes.
MATERIALS AND METHODS
Nematode
Four populations of A. besseyi (Table S1) used in this study were collected and identified by Plant Nematology Laboratory, South China Agricultural University. A. besseyi was preserved and cultured on excised carrot (Daucus carota) disks in Petri dishes at 25 ℃ in the dark (0-h light/24-h dark photoperiod) as described previously [30].
RNA isolation and reverse transcription
The total RNA of 12 samples, which include four mixed-stage nematodes samples from four populations (N10, S24, X8 and YQ) (each sample contained 1250 mixed-stage nematodes), six samples of single developmental stage (female, male and juvenile) from populations N10 and S24 (each sample contained 1000 nematodes), and two egg samples of populations N10 and S24 (each sample contained 3000 eggs), were extracted according to the instructions of the miRNeasy micro kit (Qiagen, USA), respectively. Each sample was replicated three times. The concentration of total RNA of each sample was determined using a NanoDrop2000c spectrophotometer (Thermo Fisher Scientific, USA) and its integrity and quality were confirmed by 1% agarose gel electrophoresis. 200 ng total RNA of each sample was reverse-transcribed into cDNA by TransScript One-Step gDNA Removal and cDNA Synthesis Super Mix Kit (Trans, China) according to the manufacturer’s protocol. All templates were stored at − 20 °C until use.
Cloning and bioinformatic analysis of the candidate reference genes
Referring to the transcriptome data (accession number: SRR4002929 and SRR4002930) generated by our laboratory, the fragments of four candidate reference genes actin, GAPDH, UBC and α-tubulin were cloned. The sequence of 18S rRNA (AY508035) was downloaded from NCBI. Specific primers were designed for cloning the fragments of the four candidate reference genes of A. besseyi by Primer Premier 5.0 (Primer Biosoft, USA) (Table S2). PCR was performed with a cDNA template of the mixed-stage nematodes of N10. The reaction procedure in a 20 μL volume that included 2.0 μL template cDNA, 200 nM of gene-specific primers and 10 μL of 2 × Taq master mix (GenStar, China) was as follows: 94 °C for 2 min; 40 cycles of 94 °C for 30 s, 58 °C for 30 s and 72 °C for 2 min; and a final elongation step at 72 °C for 5 min. The purified products were sequenced and subjected to BLAST search via the NCBI website, and ORFfinder (https://www.ncbi.nlm.nih.gov/orffinder/) was used to predict their corresponding amino acid sequences.
Primer design and RT-qPCR
The RT-qPCR primers were designed based on conserved sequence regions of the five candidate reference genes by Primer Premier 5.0 (Primer Biosoft, USA) and Primer-blast (www.ncbi.nlm.nih.gov/tools/primer-blast/) (Table S3). The RT-qPCR analysis was performed with each reaction mixture in a 20 μL volume containing 250 nM of each primer, 2 µL of diluted cDNA templates and 10 µL of AceQ qPCR SYBR® greenMaster Mix (Vazyme, China) on Bio-Rad CFX-96 Real-Time PCR system (Bio-Rad, USA). The thermal cycling conditions were as follows: 95 °C for 60 s, followed by 40 cycles of 95 °C for 15 s and 60 °C for 60 s. Melting curve analysis was performed after 40 cycles to verify the reaction specificity. Then, the RT-qPCR products were visualized by 2% agarose gel. A standard curve was created by a fivefold dilution series of the mixed cDNA from all samples, and PCR amplification efficiency (E) values of all primer pairs were calculated based on the standard curves according to the formula E = [10 (− 1/slope) − 1] × 100% [6]. Each RT-qPCR analysis was performed with three biological replicates. In addition, a non-template control was included for each replicate with RNase—free water instead of cDNA templates.
Data analysis
To homogenize the runs, the qPCR threshold used to determine threshold values (Ct) was set the same fluorescence value (RFU). The Ct value for each amplification curve was determined by the Bio-Rad CFX-96 Manager software (Bio-Rad, USA). The stability of the five candidate reference genes under the three experimental conditions, including the four life stages (female, male, juvenile and egg) of populations N10, the four life stages (femal, male, juvenile and egg) of populations S24 and the mixed-stage nematodes of all four populations were evaluated by ΔCt [20], geNorm [21], NormFinder [22] and RefFinder [23]. All raw Ct values were subjected to analysis by one-way ANOVA and tested for differences between groups at a 5% level using Turkey Test.
Results
Cloning and bioinformatic analysis of the candidate reference genes
Using the mixed cDNA of all samples as a template, the specific fragments of actin (876 bp), GAPDH (563 bp), UBC (615 bp) and α-tubulin (1104 bp) were amplified by PCR using specific primers of the four candidate reference genes (Fig. S1). The amplified products were sequenced and the results showed that the size of all sequences was consistent with expectations. The amino acid sequences encoded by the fragment of the each candidate reference gene were predicted by ORFfinder (https://www.ncbi.nlm.nih.gov/orffinder/), respectively. The nucleic acid sequences and predicted amino acid sequences of each reference gene fragment were compared with that of other nematode genes in NCBI by performing BLAST analysis. The results showed that the similarity of the nucleic acid sequences and predicted amino acid sequences between the four candidate reference genes of A. besseyi and corresponding genes of other nematodes were all greater than 70% (Table S4). Among them, the amino acid sequence similarity between actin of A. besseyi and that of Bursaphelenchus xylophilus and Caenorhabdit elegans has reached 100%. Further analysis showed that the predicted amino acid sequences of the four candidate reference genes all have conserved domains encoded by the corresponding genes (Fig. S2). Therefore, the actin, GAPDH, UBC and α-tubulin gene fragments amplified in this study were actin (KY992856), GAPDH (KY992853), UBC (KY992854) and α-tubulin (KY992855) gene fragments of A. besseyi.
Primers analysis of the candidate reference genes in RT-qPCR
The melting curves of actin, GAPDH, UBC, α-tubulin and 18S rRNA gene generated by RT-qPCR were all single peaks (Fig. S3), indicating that the RT-qPCR primers of the five candidate reference genes had good specificity and no dimer amplification. The size of the RT-qPCR products of the five candidate reference genes confirmed by 1% agarose gel electrophoresis all matched expectations (Fig. S4), which further validated the high specificity of the RT-qPCR primers.
Expression of candidate reference genes
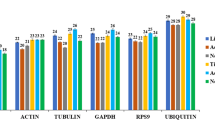
Ct value is closely related to gene expression, and the higher Ct value, the lower the gene expression. In order to compare the expression level of the five candidate reference genes, expression of these genes in 12 samples divided into three groups (the three experimental conditions), including a group consisting of mixed-stage nematodes of four populations N10, S24, X8 and YQ, a group consisting of four life stages (female, male, juvenile and egg) of population N10 and a group consisting of four life stages (female, male, juvenile and egg) of population S24 were detected by RT-qPCR. The results showed the expression level of the five candidate reference genes were not consistent across all samples (Fig. 1). The Ct values of 18S rRNA ranged from 10.22 to 14.18 in 12 samples and significantly differed among the groups (P = 0.0483). The Ct values of actin, GAPDH, UBC and α-tubulin ranged from 19.40 to 22.55, 23.8 to 26.24, 24.46 to 26.60 and 23.94 to 27.50 in all 12 samples, respectively. But there were no significant difference among the groups for the four genes (P = 0.669, P = 0.645, P = 0.207, P = 0.572). Among the five genes, 18S rRNA and α-tubulin are the genes with the highest and lowest expression level respectively, while actin, GAPDH and UBC are the genes with moderate expression level. The average Ct values of the five genes were 25.68(α-tubulin), 12.89(18S rRNA), 21.12(actin), 24.85(GAPDH) and 25.53(UBC). An ideal reference gene should have a moderate expression level with a Ct value between 15 and 30 [31, 32], the high or low expression of a reference gene will affect the accuracy of quantitative results [21]. Among the five genes, UBC showed the lowest variation in expression among the 12 samples with a difference of 2.14 between the maximum and minimum CT values. In contrast, 18S rRNA showed the highest variation in expression among the 12 samples with a difference of 3.96 between the maximum and minimum CT values, and the difference was significant among groups (P < 0.05). The results showed that the commonly used reference gene 18S rRNA of A.besseyi is not the best choice in the RT-qPCR analysis of the three experimental conditions in this study. Further analysis is still needed to screen out the optimal reference gene.
Range of all Ct values of the five candidate reference genes of Aphelenchoides besseyi. a Four life stages of N10 population. b Four life stages of S24 population. C: Four different populations. 75% of the Ct values are in the "box" range. 25% of the Ct values are in the "vertical line" range. The “short lines” represent the maximum and minimum Ct values. The "horizontal line within the square" represents the median Ct value. *P indicates significant differences between groups (P < 0.05)
Stability of reference genes
Δ Ct method analysis
The expression stability of the five candidate reference genes of A. besseyi were analyzed using the ΔCt method. The results showed that under the three experimental conditions, the expression stability of the five candidate reference genes was ordered as follows: UBC > actin > 18S rRNA > α-tubulin > GAPDH; 18S rRNA > UBC > GAPDH > actin > α-tubulin; and UBC > GAPDH > α-tubulin > actin > 18S rRNA (Table 1). Therefore, the average stability ranking of the five reference genes in descending order was UBC (1.33) > 18S rRNA (3) > actin (3.33) = GAPDH (3.33) > α-tubulin (4) (Table 1). The stability of 18S rRNA ranked first and third respectively in population S24 and population N10, while it ranked last between populations. When the data of mixed-population were excluded from analysis, the average stability ranking was UBC (1.5) > 18S rRNA (2) > actin (3) > GAPDH (4) > α-tubulin (4.5). Although the stability ranking of 18S rRNA was still lower than that of UBC, its average ranking in this case has risen by 1 place, indicating the expression level of 18S rRNA varies more between populations than within a certain population. The results showed that UBC was the most stable reference gene; 18S rRNA, actin and GAPDH were moderately stable reference genes; α-tubulin was the least stable reference gene.
geNorm analysis
The results of geNorm analysis showed that the M values of the five candidate genes were all less than 1.5 in all three experimental conditions, indicating the five genes were all evaluated as available reference genes. For the four life stages of population N10, the M values of the five candidate genes followed the order GAPDH > UBC > 18S rRNA > α-tubulin = actin (Fig. 2), indicating the stability ranking of the five genes followed the order actin = α-tubulin > 18S rRNA > UBC > GAPDH in this situation. For the four life stages of population S24, the M values of the five candidate genes followed the order α-tubulin > actin > 18S rRNA > GAPDH = UBC (Fig. 3), indicating the stability ranking of the five genes followed the order UBC = GAPDH > 18S rRNA > actin > α-tubulin in this situation. For the mixed-stage nematodes of the four populations, the M values of the five candidate genes followed the order 18S rRNA > actin > α-tubulin > GAPDH = UBC (Fig. 4), indicating the stability ranking of the five genes followed the order UBC = GAPDH > α-tubulin > actin > 18S rRNA in this situation. Therefore, the average stability ranking of the five reference genes in descending order was UBC (2) > GAPDH (2.33) > actin (3) = α-tubulin (3) > 18S rRNA(3.67). Similar to Δ Ct method results, the stability of 18S rRNA between populations was lower than that within populations, 18S rRNA was the third stable gene in both population N10 and S24 as well as the least stable gene between populations. When the data of mixed-population were excluded from analysis, the average stability ranking was UBC (2.5) = actin (2.5) > 18S rRNA (3) = GAPDH (3) = α-tubulin (3). In this case, the average ranking and overall ranking of 18S rRNA among all genes rose from 3.67 to 3 and 5 to 3 respectively. Furthermore, for the four life stages of population N10, V3/4 was 0.195 (Fig. 2), indicating the optimal numbers of reference genes was three (18S rRNA, actin and α-tubulin) in this situation; for the four life stages of population S24, V2/3 was 0.185 (Fig. 3), indicating the optimal numbers of reference genes was two (GAPDH and UBC) in this situation; for the mixed-stage nematodes of four populations, V2/3 was 0.088 (Fig. 4), indicating the optimal numbers of reference genes was two (UBC and GAPDH) in this situation.
NormFinder analysis
The results of NormFinder analysis showed that under the three experimental conditions, the expression stability of the five candidate genes followed the order UBC > 18S rRNA > actin > α-tubulin > GAPDH, UBC > 18S rRNA > GAPDH > actin > α-tubulin, UBC > GAPDH > α-tubulin > actin > 18S rRNA, respectively (Table 2). Therefore, the average stability ranking of the five reference genes in descending order was UBC (1) > 18S rRNA (3) > GAPDH (3.33) > actin (3.67) > a-tubulin (4) (Table 2). Similarly, the stability of 18S rRNA between populations was lower than that within populations, 18S rRNA ranked second in both population N10 and S24 but ranked last between populations in terms of stability. When the data of mixed-population were excluded from analysis, the average stability ranking was UBC (1) > 18S rRNA (2) > actin (3.5) > GAPDH (4) > α-tubulin (4.5). Although 18S rRNA’s overall ranking among genes remained the same, its average ranking rose from 3 to 2 in this case. These results showed that UBC was the most stable candidate reference gene; 18S rRNA, GAPDH and actin were moderately stable reference genes, and a-tubulin was the least stable candidate reference gene under the three experimental conditions.
RefFinder analysis
The stable values of the five reference genes obtained by RefFinder analysis showed that under the three experimental conditions, the expression stability of the five reference genes followed the order UBC > actin > 18S rRNA > α-tubulin > GAPDH, UBC > 18S rRNA > GAPDH > actin > α-tubulin, UBC > GAPDH > α-tubulin > actin > 18S rRNA, respectively (Table 3). Therefore, the average stability ranking of the five reference genes in descending order was UBC (1) > 18S rRNA (3.33) = actin (3.33) = GAPDH (3.33) > a-tubulin (4) (Table 3). Not surprisingly, although 18S rRNA ranked second and third in population S24 and N10 respectively, it was rated as the least stable gene between populations again. When the data of mixed-population were excluded from analysis, the average stability ranking was UBC (1) > 18S rRNA (2.5) > actin (3) > GAPDH (4) > α-tubulin (4.5), although it did not affect the overall ranking of each gene, the average ranking of 18S rRNA rose from 3.33 to 2.5 in this case. According to the ranking, we can confirm that UBC is the most stable candidate reference genes; 18S rRNA, actin and GAPDH are moderately stable reference genes, and a- tubulin is the least stable reference gene.
Discussion
qPCR has been commonly used for gene expression pattern analysis due to its high speed, sensitivity and specificity. However, as the technology is applied more and more widely, its shortcomings have also been gradually understood [33]. Due to its high sensitivity, the relative expression level of target genes may be significantly affected by the choice of wrong reference genes. The employment of an unstable expressed reference gene might result in wrong and invalid conclusions. Thus, it is crucial to select appropriate reference genes for stable and reproducible gene expression measurement in RT-qPCR experiments. At present, 18S rRNA is the most commonly used reference gene for A. besseyi. However, the results of this study indicated that 18S rRNA wasn’t the optimal reference gene among the five candidates.
In this study, the four candidate reference genes, actin, GAPDH, UBC, and a-tubulin which have been extensively researched in other species were cloned for the first time from A. besseyi. Actin is an important cytoskeleton protein involved in the process of cell secretion, phagocytosis, migration, cytoplasmic flow and cytoplasmic separation (Fu et al. [34]). GAPDH is an enzyme involved in the formation of ATP in the glycolysis reaction and is expressed in almost all tissues [35]. UBC is involved in the basic biochemical metabolism of organisms and is an important protein degradation enzyme [36]. a-tubulin is a microtubule-like protein that plays an indispensable role in the maintenance of cell shape, motility and intracellular material transport [37]. These genes are all basic components of organelles and necessary for the maintenance of life activities of the organism or participate in basic biochemical metabolic processes. In theory, they can be stably expressed in all cells and physiological conditions, suggesting that they should be ideal reference genes. In this study, we cloned actin, GAPDH, UBC and α-tubulin fragments based on the data from the transcriptome database of A. besseyi. The expression levels of these four genes and 18S rRNA under three experimental conditions were determined by RT-qPCR and their stabilities were analyzed by ΔCt, geNorm, NormFinder and RefFinder. The analysis results of ΔCt indicated that UBC was the most suitable candidate reference gene while α-tubulin was the least suitable reference gene; The analysis results of geNorm indicated that UBC was the most suitable candidate reference gene while 18S rRNA was the least suitable reference gene; The analysis results of NormFinder indicated that UBC was the most suitable candidate reference gene while a-tubulin was the least suitable reference gene; The analysis results of RefFinder indicated that UBC was the most suitable candidate reference gene while a-tubulin was the least suitable reference gene. The results of different softwares used to analyze the stability of the candidate genes were inconsistent because the algorithm of each softwares was different. Therefore, the use of multiple softwares for screening the reference genes can avoid the bias caused by the use of single software, and it is necessary to comprehensively consider the results of various analysis methods to select the optimal reference gene. Although the algorithms of the four methods differ, they all calculated that UBC was the most suitable candidate reference gene under the three experimental conditions, suggesting that UBC could be an ideal reference gene in RT-qPCR experiments for A. besseyi. The analysis results of three softwares including ΔCt, NormFinder and RefFinder deduced that 18S rRNA was the second best candidate reference gene, whereas geNorm deduced it as the least suitable candidate reference gene.
Interestingly, the results of all four analysis softwares deduced that 18S rRNA was the least stable reference gene under the experimental conditions of mixed-stage nematodes of different populations. The removal of the mixed-population data resulted in an average ranking increase of 1, 0.67, 1 and 0.83 in the four software of 18S rRNA, respectively, suggesting the expression stability of 18S rRNA between populations is not as good as it is within populations. It has been reported that ribosomal subunit copy number of A. besseyi have differences and intra-individual variation [18, 19], implying that variable results are easily produced in any experiment which includes mixed populations and that any ribosomal target has limited use when studying between species or even between populations of nematodes where the copy number of the ribosomal subunit per individual has not been previously established. By contrast, the copy number of the ribosomal subunit per individual within a population is likely to be consistent. This may well explain the results that the expression variation of 18S rRNA is low in a single population and large in different populations in this study. Therefore, although 18S rRNA is a traditional reference gene in RT-qPCR analysis for A. besseyi, it is not the optimal reference gene. The shortcomings of 18S rRNA, including high expression level, variable copy number and distinctive gene types, limit it as an ideal reference gene. Using 18S rRNA as a reference gene in RT-qPCR experiments is likely to lead to inaccurate results.
In another paper published by our group, eIF5A was tested as the best reference genes for R. similis [10]. Both papers analyzed the expression stability of 18 s rRNA, actin, and α-tubilin in two nematode species. The difference between them was that in the previous paper the expression stability of Rps21, eIF5A, UBI, and β-PP1 in R. similis were analyzed while the expression stability of GAPDH and UBC in A. besseyi were analyzed in this paper. Some different candidate reference genes were chosen between these two papers because even the same reference gene can have very different expression level in different nematode species, and not all reference genes are suitable for different nematode species, therefore, cloning and searching for more reference genes are more beneficial to the follow-up research. In the preliminary screening work of this paper, we have also selected eIF5A of A. besseyi as a candidate gene, however, the preliminary results showed that its expression level was too low in A. besseyi, and its ct values were both greater than 30 in different populations and different developmental stages (data not shown), indicating that it cannot work as an ideal reference gene. Moreover, due to the low expression of eIF5A, its ct value cannot be detected after gradient dilution, which makes its amplification curve and amplification efficiency (E value) unavailable. Based on this, we gave up further analysis of eIF5A in subsequent studies. It is worth noting that eIF5A is moderately expressed in R. similis, which was different from that of A. besseyi, this fact confirms the above idea that even the same reference gene can have quite different expression level in different nematode species.
Although we have found a better reference gene than the one commonly used—18S rRNA in this study, only five candidate reference genes were analyzed under three experimental conditions, thus, whether there is a better reference gene than UBC remains to be further studied.
Conclusions
In this study, four software programs, including ΔCt, geNorm, NormFinder and RefFinder, were used to analyze the expression stability of five candidate genes under three experimental conditions, the result revealed that UBC was an ideal reference gene in all experimental conditions, indicating UBC could be a suitable reference gene in RT-qPCR experiments for A. besseyi.
Data availability
The transcriptome database used in this study are available on Sequence Read Archive (SRA) (accession number: SRR4002929 and SRR4002930). The cDNA sequences of the five reference genes are available on GenBank: actin (KY992856), GAPDH (KY992853), UBC (KY992854), α-tubulin (KY992855) and 18 s rRNA (AY508035.1). The original data of the real-time PCR experiments are available on Supplementary material files.
Code availability
geNorm: (https://genorm.cmgg.be/). NormFinder v20:(https://www.moma.dk/normfinder-software/). ΔCt and RefFinder: (https://www.heartcure.com.au/reffinder/?type=reference).
References
Skinner DZ (2014) Real-time PCR: advanced technologies and applications. Crop Sci 54:455
Pfaffl M (2007) Relative quantification. Real-time PCR. Taylor & Francis, Milton Park, pp 89–108
Nolan T, Hands RE, Bustin SA (2006) Quantification of mRNA using real-time RT-PCR. Nat Protoc 1:1559
Quackenbush J (2002) Microarray data normalization and transformation. Nat Genet 32:496–501
Hu Y, Ni Y, Ren L, Dai J, Zhao R (2008) Leptin is involved in the effects of cysteamine on egg laying of hens, characteristics of eggs, and posthatch growth of broiler offspring. Poult Sci 87:1810–1817
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408
Hoogewijs D, Houthoofd K, Matthijssens F, Vandesompele J, Vanfleteren JR (2008) Selection and validation of a set of reliable reference genes for quantitative sod gene expression analysis in C. elegans. BMC Mol Biol 9:9
Lecová L, Růžičková M, Laing R, Vogel H, Szotáková B, Prchal L, Lamka J, Vokřál I, Skálová L, Matoušková P (2015) Reliable reference gene selection for quantitative real time PCR in Haemonchus contortus. Mol Biochem Parasitol 201:123–127
Sabeh M, Duceppe MO, St-Arnaud M, Mimee B (2018) Transcriptome-wide selection of a reliable set of reference genes for gene expression studies in potato cyst nematodes (Globodera spp.). PLoS ONE 13:e193840
Li J, Chen W, Yang S, Xu C, Huang X, Chen C, Xie H (2019) Screening of reference genes in real-time PCR for Radopholus similis. PeerJ 7:e6253
Mathur VK, Arjun L (1989) Improved culturing technique for Aphelenchoides besseyi by inducing anhydrobiosis. Indian J Nematol 1:10–13
Manzanilla-López R, Palomares-Rius J, Wesemael W (2013) Top 10 plant-parasitic nematodes in molecular plant pathology. Mol Plant Pathol 9:946–961
Cheng X, Xiang Y, Xie H, Xu CL, Xie TF, Zhang C, Li Y (2013) Molecular characterization and functions of fatty acid and retinoid binding protein Gene (Ab-far-1) in Aphelenchoides besseyi. PLoS ONE 8:e66011
Feng H, Wei L, Chen H, Zhou Y (2015) Calreticulin is required for responding to stress, foraging, and fertility in the white-tip nematode, Aphelenchoides besseyi. Exp Parasitol 155:58–67
Radonić A, Thulke S, Mackay IM, Landt O, Siegert W, Nitsche A (2004) Guideline to reference gene selection for quantitative real-time PCR. Biochem Biophys Res Commun 313:856–862
Stürzenbaum SR, Kille P (2001) Control genes in quantitative molecular biological techniques: the variability of invariance. Comp Biochem Physiol B 130:281–289
Tong Z, Gao Z, Wang F, Zhou J, Zhang Z (2009) Selection of reliable reference genes for gene expression studies in peach using real-time PCR. BMC Mol Biol 10:1–13
Buonicontro DS, Roberts DM, Oliveira CM, Blok V, Neilson R, Oliveira RDAD (2017) A rapid diagnostic for detection of Aphelenchoides besseyi and A. fujianensis based on real-time PCR. Plant Dis 102:8–17
De Jesus DS, Oliveira CMGA, Balbino HM, Mackenzie KM, Neilson R, Prior T, Roberts D, Blok V, Oliveira RDAD (2016) Morphological and molecular characterisation of Aphelenchoides besseyi and A. fujianensis (Nematoda: Aphelenchoididae) from rice and forage grass seeds in Brazil. Nematology 18:337–356
Silver N, Best S, Jiang J, Thein S (2006) Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol Biol 1:33
Vandesompele J, Preter KD, Pattyn F, Poppe B, Roy NV, Paepe AD, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3:h31–h34
Andersen C, Jensen J, Ørntoft T (2004) Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Can Res 64:5245–5250
Xie F, Xiao P, Chen D, Xu L, Zhang B (2012) miRDeepFinder: a miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol Biol 80:75–84
Ayca CC, Elif D, Serpil E, Erkan K, Duygu D, Betul K (2012) A novel strategy for selection and validation of reference genes in dynamic multidimensional experimental design in yeast. PLoS ONE 7:e38351
Vieira A, Cabral A, Fino J, Azinheira HG, Loureiro A, Talhinhas P, Pires AS, Varzea V, Moncada P, Oliveira H (2016) Comparative validation of conventional and RNA-Seq data-derived reference genes for qPCR expression studies of Colletotrichum kahawae. PLoS ONE 11:e150651
Hu Y, Xie S, Yao J (2016) Identification of novel reference genes suitable for qRT-PCR normalization with respect to the zebrafish developmental stage. PLoS ONE 11:e149277
Zhan C, Zhang Y, Ma J, Wang L, Jiang W, Shi Y, Wang Q (2014) Identification of reference genes for qRT-PCR in human lung squamous-cell carcinoma by RNA-Seq. Acta Biochim Biophys Sin 46:330
Chang E, Shi S, Liu J, Cheng T, Xue L, Yang X, Yang W, Lan Q, Jiang Z (2012) Selection of reference genes for quantitative gene expression studies in Platycladus orientalis (Cupressaceae) using real-time PCR. PLoS ONE 7:e33278
Wang D, Xu C, Ding S, Huang X, Cheng X, Zhang C, Chen C, Xie H (2018) Identification and function of FAR protein family genes from a transcriptome analysis of Aphelenchoides besseyi. Bioinformatics 34:2936–2943
Pei YY, Cheng X, Chun-Ling XU, Yang ZF, Xie H (2012) Virulence of part populations of Aphelenchoides besseyi on rice in China. Chin J Rice Sci 46:522–529
Lilly ST, Drummond RSM, Pearson MN, Macdiarmid RM (2011) Identification and validation of reference genes for normalization of transcripts from virus-infected Arabidopsis thaliana. Mol Plant Microbe Interact 24:294–304
Wan H, Zhao Z, Qian C, Sui Y, Malik AA, Chen J (2010) Selection of appropriate reference genes for gene expression studies by quantitative real-time polymerase chain reaction in cucumber. Anal Biochem 399:257–261
Bustin S (2002) Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. J Mol Endocrinol 29:23–39
Fu G, Yang T, Li W, Wang J (2013) Cloning and sequence analysis of actin gene fragment from Iris lactea var.chinensis Fisch.Koidz. J Northeast Agric Univ (English Ed) 20:12–16
Barber RD, Dan WH, Coleman RA, Clark BJ (2005) GAPDH as a housekeeping gene: analysis of GAPDH mRNA expression in a panel of 72 human tissues. Physiol Genomics 21:389
Deng L, Wang C, Spencer E, Yang L, Braun A, You J, Slaughter C, Pickart C, Chen ZJ (2000) Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell 103:351–361
Wolf KW, Spanel-Borowski K (2013) Acetylation of α-tubilin in different bovine cell types: implications for microtubule dynamics in interphase and mitosis. Cell Biol Int 19:43–52
Acknowledgements
This worked was funded by National Natural Science Foundation of China (Nos. 31871939 and 31371920).
Funding
This worked was funded by National Natural Science Foundation of China (No. 31871939 and 31371920).
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments: ZXZ HX. Performed the experiments: JYL ZXZ. Analyzed the data: JYL ZXZ. Contributed reagents/materials/analysis tools: DDW CLX ML. Wrote the paper: JYL ZXZ HX.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Li, J., Zhang, Z., Xu, C. et al. Identification and validation of reference genes for real-time RT-PCR in Aphelenchoides besseyi. Mol Biol Rep 47, 4485–4494 (2020). https://doi.org/10.1007/s11033-020-05547-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-020-05547-8