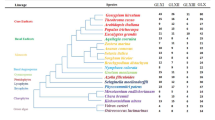
Abstract
The multidrug and toxic compound extrusion (MATE) protein belongs to a secondary transporter family, which plays a role in transporting different kinds of substrates like phytohormones and secondary metabolites. In plant, MATE transporters related to the endogenous and exogenous mechanisms of detoxification for secondary metabolites such as alkaloids, flavonoids, anthocyanins and other secondary metabolites have been studied. However, a genome-wide analysis of the MATE family is rarely reported in upland cotton (Gossypium hirsutum L.). In the study, a total of 72 GhMATEs were identified from the genome of upland cotton, which were classified into four subfamilies with possible diverse functions such as transport of proanthocyanidins (PAs), accumulation of alkaloids, extrusion of xenobiotic compounds, regulation of disease resistance and response to abiotic stresses. Meanwhile, the gene structure, evolutionary relationship, physical location, conservative motifs, subcellular localization and gene expression pattern of GhMATEs have been further analysed. Three of these MATE genes (GhMATE12, GhMATE16 and GhMATE38) were identified as candidate genes due to their functions in transport of PA similar to GhTT12. These results provide a new perspective on upland cotton MATE gene family for their potential roles in transport of PA and a theoretical basis for further analyzing the function of MATE genes and improving the fiber quality of brown cotton.









Similar content being viewed by others
References
Saier MH Jr, Paulsen IT (2001) Phylogeny of multidrug transporters. Semin Cell Dev Biol 12:205–213
Omote H, Hiasa M, Matsumoto T, Otsuka M, Moriyama Y (2006) The MATE proteins as fundamental transporters of metabolic and xenobiotic organic cations. Trends Pharmacol Sci 27:587–593
Magalhaes JV (2010) How a microbial drug transporter became essential for crop cultivation on acid soils: aluminium tolerance conferred by the multidrug and toxic compound extrusion (MATE) family. Ann Bot 106:199–203
He X, Szewczyk P, Karyakin A, Evin M, Hong W-X, Zhang Q, Chang G (2010) Structure of a cation-bound multidrug and toxic compound extrusion transporter. Nature 467:991–994
Wang L, Bei X, Gao J, Li Y, Yan Y, Hu Y (2016) The similar and different evolutionary trends of MATE family occurred between rice and Arabidopsis thaliana. BMC Plant Biol 16:207
Zhu H, Wu J, Jiang Y, Jin J, Zhou W, Wang Y, Han G, Zhao Y, Cheng B (2016) Genomewide analysis of MATE-type gene family in maize reveals microsynteny and their expression patterns under aluminum treatment. J Genet 95:691–704
Liu J, Li Y, Wang W, Gai J, Li Y (2016) Genome-wide analysis of MATE transporters and expression patterns of a subgroup of MATE genes in response to aluminum toxicity in soybean. BMC Genomics 17:223
Chen L, Liu Y, Liu H, Kang L, Geng J, Gai Y, Ding Y, Sun H, Li Y (2015) Identification and expression analysis of MATE genes involved in flavonoid transport in blueberry plants. PLoS ONE 10:e0118578
Yazaki K, Sugiyama A, Morita M, Shitan N (2008) Secondary transport as an efficient membrane transport mechanism for plant secondary metabolites. Phytochem Rev 7:513–524
Santos ALD, Chaves-Silva S, Yang L, Maia LGS, Chalfun-Júnior A, Sinharoy S, Zhao J, Benedito VA (2017) Global analysis of the MATE gene family of metabolite transporters in tomato. BMC Plant Biol 17:185
Lu P, Magwanga RO, Guo X, Kirungu JN, Lu H, Cai X, Zhou Z, Wei Y, Wang X, Zhang Z, Peng R, Wang K, Liu F (2018) Genome-wide analysis of multidrug and toxic compound extrusion (MATE) family in Gossypium raimondii and Gossypium arboreum and its expression analysis under salt, cadmium, and drought stress. G3 8:2483–2500
Gao J-S, Wu N, Shen Z-L, Lv K, Qian S-H, Guo N, Sun X, Cai Y-P, Lin Y (2016) Molecular cloning, expression analysis and subcellular localization of a Transparent Testa 12 ortholog in brown cotton (Gossypium hirsutum L.). Gene 576:763–769
Zhao X, Wang X (2005) Composition analysis of pigment in colored cotton fiber. Acta Agron Sin 31:456–462
Zhan S, Lin Y, Cai Y, Li Z (2007) Preliminary deductions of the chemical structure of the pigment brown in cotton fiber. Chin Bull Bot 24:99–104
Dixon RA, Liu C, Jun JH (2013) Metabolic engineering of anthocyanins and condensed tannins in plants. Curr Opin Biotechnol 24:329–335
Dixon RA, Xie DY, Sharma SB (2005) Proanthocyanidins—a final frontier in flavonoid research? New Phytol 165:9–28
Lepiniec L, Debeaujon I, Routaboul J-M, Baudry A, Pourcel L, Nesi N, Caboche M (2006) Genetics and biochemistry of seed flavonoids. Annu Rev Plant Biol 57:405–430
Ariga T, Asao Y, Sugimoto H, Yokotsuka T (1981) Occurrence of astringent oligomeric proanthocyanidins in legume seeds. Agric Biol Chem 45:2705–2708
Gabetta B, Fuzzati N, Griffini A, Lolla E, Pace R, Ruffilli T, Peterlongo F (2000) Characterization of proanthocyanidins from grape seeds. Fitoterapia 71:162–175
Gu L, Kelm MA, Hammerstone JF et al (2004) Concentrations of proanthocyanidins in common foods and estimations of normal consumption. J Nutr 134:613–617
Santos-Buelga C, Scalbert A (2000) Proanthocyanidins and tannin-like compounds–nature, occurrence, dietary intake and effects on nutrition and health. J Sci Food Agric 80:1094–1117
Serafini M, Bugianesi R, Maiani G, Valtuena S, De Santis S, Crozier A (2003) Plasma antioxidants from chocolate. Nature 424:1013
Zhao J, Dixon RA (2009) MATE transporters facilitate vacuolar uptake of epicatechin 3′-O-glucoside for proanthocyanidin biosynthesis in Medicago truncatula and Arabidopsis. Plant Cell 21:2323–2340
Zhao J, Huhman D, Shadle G, He X-Z, Sumner LW, Tang Y, Dixon RA (2011) MATE2 mediates vacuolar sequestration of flavonoid glycosides and glycoside malonates in Medicago truncatula. Plant Cell 23:1536–1555
Paterson AH, Wendel JF, Gundlach H et al (2012) Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 492:423–427
Wang K, Wang Z, Li F et al (2012) The draft genome of a diploid cotton Gossypium raimondii. Nat Genet 44:1098–1103
Li F, Fan G, Wang K et al (2014) Genome sequence of the cultivated cotton Gossypium arboreum. Nat Genet 46:567–572
Eddy SR (1996) Hidden Markov models. Curr Opin Struct Biol 6:361–365
Finn RD, Mistry J, Schuster-Böckler B et al (2006) Pfam: clans, web tools and services. Nucleic Acids Res 34:247–251
Letunic I, Doerks T, Bork P (2012) SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res 40:302–305
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A (2003) ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res 31:3784–3788
Zhang C, Zhang H, Zhao Y, Jiang H, Zhu S, Cheng B, Xiang Y (2013) Genome-wide analysis of the CCCH zinc finger gene family in Medicago truncatula. Plant Cell Rep 32:1543–1555
Hu B, Jin J, Guo A-Y, Zhang H, Luo J, Gao G (2014) GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31:1296–1297
Bailey TL, Elkan C (1995) The value of prior knowledge in discovering motifs with MEME. Proc Int Conf Intell Syst Mol Biol 3:21–29
He H, Dong Q, Shao Y, Jiang H, Zhu S, Cheng B, Xiang Y (2012) Genome-wide survey and characterization of the WRKY gene family in Populus trichocarpa. Plant Cell Pep 31:1199–1217
Wang Y, Tang H, Debarry JD et al (2012) MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res 40:e49
Wang F, Dong Q, Jiang H, Zhu S, Chen B, Xiang Y (2012) Genome-wide analysis of the heat shock transcription factors in Populus trichocarpa and Medicago truncatula. Mol Biol Rep 39:1877–1886
Li L, Stoeckert CJ, Roos DS (2003) OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res 13:2178–2189
Krzywinski M, Schein J, Birol I et al (2009) Circos: an information aesthetic for comparative genomics. Genome Res 19:1639–1645
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆CT method. Methods 25:402–408
Ikegami A, Akagi T, Potter D, Yamada M, Sato A, Yonemori K, Kitajima A, Inoue K (2009) Molecular identification of 1-Cys peroxiredoxin and anthocyanidin/flavonol 3-O-galactosyltransferase from proanthocyanidin-rich young fruits of persimmon (Diospyros kaki Thunb.). Planta 230:841–855
Li Y, Tanner G, Larkin PJ (1996) The DMACA-HCl protocol and the threshold proanthocyanidin content for bloat safety in forage legumes. J Sci Food Agric 70:89–101
Li L, He Z, Pandey GK, Tsuchiya T, Luan S (2002) Functional cloning and characterization of a plant efflux carrier for multidrug and heavy metal detoxification. J Biol Chem 277:5360–5368
Tiwari M, Sharma D, Singh M, Tripathi RD, Trivedi PK (2014) Expression of OsMATE1 and OsMATE2 alters development, stress responses and pathogen susceptibility in Arabidopsis. Sci Rep 4:3964
Otto SP (2007) The evolutionary consequences of polyploidy. Cell 131:452–462
Soltis PS, Soltis DE (2009) The role of hybridization in plant speciation. Annu Rev Plant Biol 60:561–588
Brenchley R et al (2012) Analysis of the bread wheat genome using whole-genome shotgun sequencing. Nature 491:705–710
Moore RC, Purugganan MD (2003) The early stages of duplicate gene evolution. Proc Natl Acad Sci USA 100:15682–15687
Cannon SB, Mitra A, Baumgarten A, Young ND, May G (2004) The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol 4:10
Thompson EP, Wilkins C, Demidchik V, Davies JM, Glover BJ (2009) An Arabidopsis flavonoid transporter is required for anther dehiscence and pollen development. J Exp Bot 61:439–451
Gomez C, Terrier N, Torregrosa L et al (2009) Grapevine MATE-type proteins act as vacuolar H+-dependent acylated anthocyanin transporters. Plant Physiol 150:402–415
Gomez C, Conejero G, Torregrosa L, Cheynier V, Terrier N, Ageorges A (2011) In vivo grapevine anthocyanin transport involves vesicle-mediated trafficking and the contribution of anthoMATE transporters and GST. Plant J 67:960–970
Shoji T, Inai K, Yazaki Y et al (2009) Multidrug and toxic compound extrusion-type transporters implicated in vacuolar sequestration of nicotine in tobacco roots. Plant Physiol 149:708–718
Pérez-Díaz R, Ryngajllo M, Pérez-Díaz J, Peña-Cortés H, Casaretto JA, González-Villanueva E, Ruiz-Lara S (2014) VvMATE1 and VvMATE2 encode putative proanthocyanidin transporters expressed during berry development in Vitis vinifera L. Plant Cell Rep 33:1147–1159
Morita M, Shitan N, Sawada K et al (2009) Vacuolar transport of nicotine is mediated by a multidrug and toxic compound extrusion (MATE) transporter in Nicotiana tabacum. Proc Natl Acad Sci USA 106:2447–2452
Mathews H, Clendennen SK, Caldwell CG et al (2003) Activation tagging in tomato identifies a transcriptional regulator of anthocyanin biosynthesis, modification, and transport. Plant Cell 15:1689–1703
Sun X, Gilroy EM, Chini A et al (2011) ADS1 encodes a MATE-transporter that negatively regulates plant disease resistance. New Phytol 192:471–482
Wang R, Liu X, Liang S et al (2015) A subgroup of MATE transporter genes regulates hypocotyl cell elongation in Arabidopsis. J Exp Bot 66:6327–6343
Rogozin IB, Wolf YI, Sorokin AV, Mirkin BG, Koonin EV (2003) Remarkable interkingdom conservation of intron positions and massive, lineage-specific intron loss and gain in eukaryotic evolution. Curr Biol 13:1512–1517
Acknowledgements
We thank Dr. Da-Hui Li for assisting the experimental technology and revising the original manuscript, and are thankful for Prof. Zhao-Rong Deng to offer the biochemical reagents. This work was sponsored by State Key Laboratory of Cotton Biology Open Fund (No. CB2018A01 to J.-S. G.) and the National Natural Science Foundation of China (Grant Nos. 31672497 to J.-S. G.; 31572468 to Y. M.). It was also supported by the National Key Research and Development Program of China (No. 2016YFD0300205-3 to Z.-R. D.).
Author information
Authors and Affiliations
Contributions
J-SG, YL and YM conceived the idea, LX, WC, G-YS and Z-LS analysed the data, J-SG, LX and Z-LS wrote the manuscript, NG, Y-PC and XS assisted in sequence analysis. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplement Fig. S1
Chromosomal location and gene duplication of 72 MATE genes on 20 chromosomes of upland cotton. The name of chromosome is indicated at the top of each bar. The scale on the left is in Mb. The MATE gene on the left side of each chromosome corresponds to the approximate location on each chromosome. The segmentally duplicated genes are connected by dashed lines. (TIF 4285 KB)
Rights and permissions
About this article
Cite this article
Xu, L., Shen, ZL., Chen, W. et al. Phylogenetic analysis of upland cotton MATE gene family reveals a conserved subfamily involved in transport of proanthocyanidins. Mol Biol Rep 46, 161–175 (2019). https://doi.org/10.1007/s11033-018-4457-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-018-4457-4