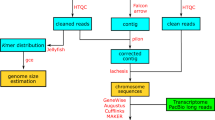
Abstract
Red swamp crayfish, Procambarus clarkii, presently is an important aquatic commercial species in China. The crayfish is a hot area of research focus, and its genetic improvement is quite urgent for the crayfish aquaculture in China. However, the knowledge of its genomic landscape is limited. In this study, a survey of P. clarkii genome was investigated based on Illumina’s Solexa sequencing platform. Meanwhile, its genome size was estimated using flow cytometry. Interestingly, the genome size estimated is about 8.50 Gb by flow cytometry and 1.86 Gb with genome survey sequencing. Based on the assembled genome sequences, total of 136,962 genes and 152,268 exons were predicted, and the predicted genes ranged from 150 to 12,807 bp in length. The survey sequences could help accelerate the progress of gene discovery involved in genetic diversity and evolutionary analysis, even though it could not successfully applied for estimation of P. clarkii genome size.




Similar content being viewed by others
References
Yue GH, Li J, Bai Z, Wang CM, Feng F (2010) Genetic diversity and population structure of the invasive alien red swamp crayfish. Biol Invasions 12:2697–2706
Li YH, Guo XW, Cao XJ et al (2012) Population genetic structure and post-establishment dispersal patterns of the red swamp crayfish Procambarus clarkii in China. PLoS ONE 7(7):e40652. https://doi.org/10.1371/journal.pone.0040652
Li YH, Guo XW, Chen LP et al (2015) Inferring invasion history of red swamp crayfish (Procambarus clarkii) in China from mitochondrial control region and nuclear intron sequences. Int J Mol Sci 16(7):14623–14639. https://doi.org/10.3390/ijms160714623
Jiang HC, Xing ZJ, Lu W et al (2014) Transcriptome analysis of red swamp crawfish Procambarus clarkii reveals genes involved in gonadal development. PLoS ONE 9(8):e105122. https://doi.org/10.1371/journal.pone.0105122
Huner JV (1988) Procambarus in North America and elsewhere. In: Holdich DM, Lowery RS (eds) Freshwater crayfish: biology, management and exploitation. Timber Press, Portland, pp. 239–261
Zhu ZY, Yue GH (2008) Eleven polymorphic microsatellites isolated from red swamp crayfish, Procambarus clarkii. Mol Ecol Resour 8:796–798
Shen HS, Hu YC, Ma YC et al (2014) In-Depth transcriptome analysis of the red swamp crayfish Procambarus clarkii. PLoS ONE 9(10):e110548. https://doi.org/10.1371/journal.pone.0110548
Alfei L, Cavallo D, Eleuteri P et al (1996) Nuclear DNA content in Salmo fibreni in Lake Posta Fibreno, Italy. J Fish Biol 48(6):1051–1058
Fafanđel M, Bihari N, Smodlaka M, Ravlic S (2008) Hemocytes/coelomocytes DNA content in five marine invertebrates: cell cycles and genome sizes. Biologia 63(5):730–736
Juchno D, Lackowska B, Boron A, Kilarski W (2010) DNA content of hepatocyte and erythrocyte nuclei of the spined loach (Cobitis tania L.) and its polyploidy froms. Fish Physiol Biochem 36(3):523–529
Liu L, Cui ZX, Song CW et al (2016) Flow cytometric analysis of DNA content for four commercially important crabs in china. Acta Oceanol Sin 35(6):7–11
Lu M, An H, Li L (2016) Genome survey sequencing for the characterization of the genetic background of Rosa roxburghii Tratt and leaf ascorbate metabolism genes. PLoS ONE 11(2):e0147530. https://doi.org/10.1371/journal.pone.0147530
Yi S, Li Y, Shi L, Zhang L et al (2018) Characterization of Population Genetic Structure of red swamp crayfish, Procambarus clarkii, in China. Sci Rep. https://doi.org/10.1038/s41598-018-23986-z
Dai M, Thompson RC, Maher C et al (2010) NGSQC: cross-platform quality analysis pipeline for deep sequencing data. BMC Genom. https://doi.org/10.1186/1471-2164-11-S4-S7
Luo RB, Liu BH, Xie YL et al (2012) SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience 1(1):18
Zhang GF, Fang XD, Guo XM et al (2012) The oyster genome reveals stress adaptation and complexity of shell formation. Nature 490(7418):49–54
Kim EB, Fang X, Fushan AA et al (2011) Genome sequencing reveals insights into physiology and longevity of the naked mole rat. Nature 479(7372):223–227
Marcais G, Kingsford C (2011) A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 27:764–770
Liu BH, Shi YJ, Yuan JY et al (2013) Estimation of genomic characteristics by analyzing k-mer frequency in de novo genome projects. Quant Biol 35(s1–3):62–67
Cimino MC (1974) The nuclear DNA content of diploid and triploid Poeciliopsis and other poeciliid fishes with reference to the evolution of unisexual forms. Chromosoma 47(3):297–307
Jimenez AG, Kinsey ST, Dillaman RM, Kapraun DF (2010) Nuclear DNA content variation associated with muscle fiber hypertrophic growth in decapod crustaceans. Genome 53(3):161–171
Huang H, Tong Y, Zhang QJ, Gao LZ (2013) Genome size variation among and within camellia species by using flow cytometric analysis. PLoS ONE 8(5):e64981
Doležel J, Bartoš J, Voglmayr H, Greilhuber J (2003) Nuclear DNA content and genome size of trout and human. Cytometry A 51:127–128
Doležel J, Bartoš J (2005) Plant DNA flow cytometry and estimation of nuclear genome size. Ann Bot 95:99–110
Excoffier L, Laval G, Schneider S (2005) Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinform Online 1:47–50
Majoros WH, Pertea M, Salzberg SL (2004) TigrScan and GlimmerHMM: two open-source ab initio eukaryotic gene-finders. Bioinformatics 20:2878–2879
Wright S (1978) Evolution and the genetics of populations, v.4: variability within and among natural populations. The University of Chicago Press, Chicago
Zhu DM, Song W, Yang K et al (2012) Flow cytometric determination of genome size for eight commercially important fish species in China. In Vitro Cell Dev 48(8):507–517
Filipiak M, Tylko G, Kilarski W (2012) Flow cytometric determination of genome size in European sunbleak Leucaspius delieatus (Heckel 1838). Fish Physiol Biochem 43(2):355–362
Bachmann K, Rheinsmith EL (1973) Nuclear DNA amounts in Pacific Crustacea. Chromosoma 43(3):225–236
Ding J, Chang YQ, Wang ZC et al (2003) Analysis of DNA relative content and cell cycle of different tissues of Crassostrea gigas. Adv Mar Sci 21(2):203–208
Ding J, Chang YQ, Xing RL, Song J, Fu Q (2003) Analysis of cell cycle and nuclear DNA relative contents in different tissues using flow cytometry. J Dalian Fish U 18(3):200–203
Liu HY, Yang S, Yan HW et al (2016) Chromosome karyotype and nuclear DNA content of mantis shrimp Oratosquilla oratoria. J Dalian Ocean U 31(1):1–6
Cavallini A, Natali L (1991) Intraspecific variation of nuclear DNA content in plant species. Caryologia 44(1):93–107
Aird D, Ross MG, Chen WS et al (2011) Analyzing and minimizing PCR amplification bias in Illumina sequencing libraries. Genome Biol 12:R18. https://doi.org/10.1186/gb-2011-12-2-r18
Kajitani R, Toshimoto K, Noguchi H et al (2014) Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Res 24:1384–1395
Claros MG, Bautista R, Guerrero-Fernández D et al (2012) Why assembling plant genome sequences is so challenging. Biology 1:439–459. https://doi.org/10.3390/biology1020439
Hamilton JP, Buell CR (2012) Advances in plant genome sequencing. Plant J 70:177–190
Huang Y, Li T, Jin M, Yin S, Hui KM, Ren Q (2017) Newly identified PcToll4 regulates antimicrobial peptide expression in intestine of red swamp crayfish Procambarus clarkii. Gene 610:140–147
Yi S, Li Y, Shi L, Zhang L (2017) Novel insights into antiviral gene regulation of red swamp crayfish, Procambarus clarkii, infected with white spot syndrome virus. Genes 8(11):320
Dearborn RE, Jr Szaro BG, Lnenicka GA (1999) Cloning and characterization of AASPs: novel axon-associated SH3 binding-like proteins. J Neurobiol 38(4):581–594
Jiang B, Lou Q, Wu Z et al (2011) Retrotransposon- and microsatellite sequence-associated genomic changes in early generations of a newly synthesized allotetraploid Cucumis × hytivus Chen & Kirkbride. Plant Mol Biol https://doi.org/10.1007/s11103-011-9804-y
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 31501858) and the Fundamental Research Funds for the Central Universities (No. 2662016QD009). The study was also financed by the 2nd batch of Modern Agroindustry Technology Research System of Hubei Province. We thank Miss Xiaoran Song and Ruijing Gen, College of fisheries, Huazhong Agriculture University, for their assistance with technique support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interest.
Ethical approval
This study was conducted with ethical approval by the Institutional Animal Care and Use Committee (IACUC) of Huazhong Agricultural University (Wuhan, China) according to the national and international guidelines.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Shi, L., Yi, S. & Li, Y. Genome survey sequencing of red swamp crayfish Procambarus clarkii. Mol Biol Rep 45, 799–806 (2018). https://doi.org/10.1007/s11033-018-4219-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-018-4219-3