Abstract
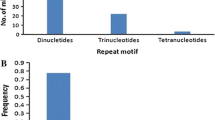
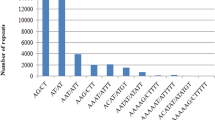
In recent years, the increased availability of the DNA sequences has given the possibility to develop and explore the expressed sequence tags (ESTs) derived SSR markers. In the present study, a total of 1956 ESTs of finger millet were used to find the microsatellite type, distribution, frequency and developed a total of 545 primer pairs from the ESTs of finger millet. Thirty-two EST sequences had more than two microsatellites and 1357 sequences did not have any SSR repeats. The most frequent type of repeats was trimeric motif, however the second place was occupied by dimeric motif followed by tetra-, hexa- and penta repeat motifs. The most common dimer repeat motif was GA and in case of trimeric SSRs, it was CGG. The EST sequences of NBS-LRR region of finger millet and rice showed higher synteny and were found on nearly same positions on the rice chromosome map. A total of eight, out of 15 EST based SSR primers were polymorphic among the selected resistant and susceptible finger millet genotypes. The primer FMBLEST5 could able to differentiate them into resistant and susceptible genotypes. The alleles specific to the resistant and susceptible genotypes were sequenced using the ABI 3130XL genetic analyzer and found similarity to NBS–LRR regions of rice and finger millet and contained the characteristic kinase-2 and kinase 3a motifs of plant R-genes belonged to NBS–LRR region. The In-silico and comparative analysis showed that the genes responsible for blast resistance can be identified, mapped and further introgressed through molecular breeding approaches for enhancing the blast resistance in finger millet.




Similar content being viewed by others
References
Latha MA, Venkateswara Rao K, Dashavantha Reddy V (2005) Production of transgenic plants resistant to leaf blast disease in finger millet (Eleusine coracana (L.) Gaertn.). Plant Sci 169:657–667
Chennaveeraiah MS, Hiremath SC (1991) Cytogenetics of minor millets. In: Tsuchiya T, Gupta PK (eds) Chromosome engineering in plants genetics breeding and evolution. Elsevier, Amsterdam, pp 613–627
Ceasar SA, Ignacimuthu S (2008) Efficient somatic embryogenesis and plant regeneration from shoot apex explants of different Indian genotypes of finger millet (Eleusine coracana (L.) Gaertn.). In Vitro Cell Dev Biol Plant 44:427–435
Zane L, Bargelloni L, Patarnello T (2002) Strategies for microsatellite isolation: a review. Mol Ecol 11:1–16
Squirrell J, Hollingsworth PM, Woodhead M, Russell J, Lowe AJ, Gibby M, Powell W (2003) How much effort is required to isolate nuclear microsatellites from plants? Mol Ecol 12:1339–1348
Weising K, Nybom H, Wolff K, Kahl G (2005) Application of DNA fingerprinting in plant sciences. DNA Fingerprinting in plants-principles, methods, and applications. CRC Press, Boca Raton, pp 235–276
Chen C, Zhou P, Choi YA, Huang S, Gmitter FG Jr (2006) Mining and characterizing microsatellites from citrus ESTs. Theor Appl Genet 112:1248–1257
Poncet V, Rondeau M, Tranchant C, Cayrel A, Hamon S, de Kochko A, Hamon P (2006) SSR mining in coffee tree EST databases: potential use of EST-SSRs as markers for the Coffea genus. Mol Gen Genomics 276:436–449
Yu J-K, Dake TM, Singh S, Benscher D, Li W, Gill B, Sorrells ME (2004) Development and mapping of EST-derived simple sequence repeat markers for hexaploid wheat. Genome 47:805–818
Mishra RK, Gangadhar BH, Jae Woong Yu, Doo Hwan Kim, Se Won Park (2011) Development and characterization of EST based SSR markers in Madagascar periwinkle (Catharanthus roseus) and their transferability in other medicinal plants. Plant Omics J 4(3):154–162
Durand J, Catherine B, Emilie C, Frigerio JM et al (2010) A fast and cost effective approach to develop and map EST-SSR markers: oak as a case study. BMC Genom 11:570–583
Senthilvel S, Jayashree B, Mahalakshmi V, Sathish Kumar P, Nakka S, Nepolean T, Hash CT (2008) Development and mapping of Simple Sequence Repeat markers for pearl millet from data mining of Expressed Sequence Tags. BMC Plant Biol 8:119
Carine AT, Wang BB, Majesta SO, Shweta D, Zhu HY, Bruce R, Nevin DY, Steven BC (2008) Identification and characterization of nucleotide-binding site-leucine-rich repeat genes in the model plant Medicago truncatula. Plant Physiol 146:5–21
Okuyama Y, Kanzaki H, Abe A, Yoshida K, Tamiru M, Saitoh H, Fujibe T, Matsumura H, Shenton M, Galam DC, Undan J, Ito A, Sone T, Terauchi R (2011) A multifaceted genomics approach allows the isolation of the rice Pia-blast resistance gene consisting of two adjacent NBS-LRR protein genes. Plant J 66:467–479
Meyers BC, Kozik A, Griego A, Kuang H, Michelmore RW (2003) Genome-wide analysis of NBS–LRR encoding genes in Arabidopsis. Plant Cell 15:809–834
Meyers BC, Dickerman AW, Michelmore RW, Sivaramakrishnan S, Sobral BW, Young ND (1999) Plant disease resistance genes encode members of an ancient and diverse protein family within the nucleotide-binding superfamily. Plant J 20:317–332
He L, Du C, Covaleda L, Xu Z, Robinson AF, Yu JZ, Kohel RJ, Zhang HB (2004) Cloning, characterization, and evolution of the NBSLRR—encoding resistance gene analogue family in polyploidy cotton (Gossypium hirsutum L.). Mol Plant Microbe Interact 17:1234–1241
Cannon SB, Shu H, Baumgarten AM, Spangler R, May G, Cook DR, Young ND (2002) Diversity, distribution, and ancient taxonomic relationships within the TIR and non-TIR NBS-LRR resistance gene subfamilies. J Mol Evol 54:548–562
Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8:4321–4326
Maniatis T, Sambrook J, Fritsch EF (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory, New York
Martins WS, Lucas DCS, Neves KFS, Bertioli DJ (2009) WebSat—A Web Software for microsatellite marker development. Bioinformation 3(6):282–283
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser 41:95–98
Reddy BLIN, Lakshmi NM, Sivaramakrishnan S (2012) Identification and characterization of EST–SSRs in finger millet (Eleusine coracana (L.) Gaertn.). J Crop Sci Biotech 15(1):9–16
Victoria FC, da Maia Luciano C, de Oliveira Antonio C (2011) In silico comparative analysis of SSR markers in plants. BMC Plant Biol 11:15–30
Zeng S, Xiao G, Guo J, Fei Z, Xu Y, Bruce A Roe, Wang Y (2010) Development of a EST dataset and characterization of EST-SSRs in a traditional Chinese medicinal plant, Epimedium sagittatum (Sieb. Et Zucc.) maxim. BMC Genom 11:94–105
Jia XP, Shi YS, Song YC (2007) Development of EST–SSR in foxtail millet (Setaria italica) [J]. Genet Resour Crop Evol 54:233–236
Kantety RV, La Rota M, Matthews DE, Sorrells ME (2002) Data mining for simple sequence repeats in expressed sequence tags from barley, maize, rice, sorghum and wheat. Plant Mol Biol 48:501–510
Thiel T, Michalek W, Varshney RK, Graner A (2003) Exploiting EST database for the development and characterization of gen-derived SSR-markers in barley (Hordeum vulgare L.). Theor Appl Genet 106:411–422
Maia LC, Souza VQ, Kopp MM, Carvalho FIF, Oliveira AC (2009) Tandem repeat distribution of gene transcripts in three plant families. Genet Mol Biol 32:1–12
Varshney RK, Thiel T, Stein N, Langridge P, Graner A (2002) In silico analysis on frequency and distribution of microsatel lites in ESTs of some cereal species. Cell Mol Biol Lett 7:537–546
Asp T, Frei UK, Didion T, Nielsen KK, Lubberstedt T (2007) Frequency, type, and distribution of EST–SSRs from three genotypes of Lolium perenne, and their conservation across orthologous sequences of Festuca arundinacea, Brachypodium distachyon, and Oryza sativa. BMC Plant Biol 7:36–47
Srinivasachary Dida MM, Gale MD, Devos KM (2007) Comparative analyses reveal high levels of conserved colinearity between the finger millet and rice genomes. Theor Appl Genet 115:489–499
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Panwar P, Jha AK, Pandey PK, Gupta Arun K, Kumar Anil (2011) Functional markers based molecular characterization and cloning of resistance gene analogs encoding NBS-LRR disease resistance proteins in finger millet (Eleusine coracana). Mol Biol Rep 38:3427–3436
Reddy BL, Srinivas Reddy D, Lakshmi Narasu M, Sivaramakrishnan S (2011) Characterization of disease resistance gene homologues isolated from finger millet (Eleusine coracana L. Gaertn). Mol Breed 27(3):315–328
Traut TW (1994) The function and consensus motifs of nine types of peptide segments that form different types of nucleotide binding sites. Eur J Biochem 222:9–19
Morel JB, Dangl JL (1997) The hypersensitive response and the induction of cell death in plants. Cell Death Differ 4:671–683
Pan Q, Wendel J, Fluhr R (2000) Divergent evolution of plant NBS–LRR resistance gene homologues in dicot and cereal genomes. J Mol Evol 50:203–213
Jeong SC, Hayes AJ, Biyashev RM, Saghai Maroof MA (2001) Diversity and evolution of a non-TIR-NBS sequence family that clusters to a chromosomal ‘hotspot’ for disease resistance genes in soybean. Theor Appl Genet 103:406–414
Penuela S, Danesh D, Young ND (2002) Targeted isolation, sequence analysis, and physical mapping of non-TIR NBSLRR genes in soybean. Theor Appl Genet 104:261–272
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Kalyana Babu, B., Pandey, D., Agrawal, P.K. et al. In-silico mining, type and frequency analysis of genic microsatellites of finger millet (Eleusine coracana (L.) Gaertn.): a comparative genomic analysis of NBS–LRR regions of finger millet with rice. Mol Biol Rep 41, 3081–3090 (2014). https://doi.org/10.1007/s11033-014-3168-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-014-3168-8