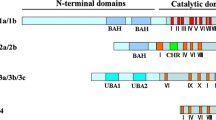
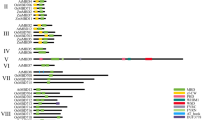
Abstract
Histone lysine methylation plays a pivotal role in a variety of developmental and physiological processes through modifying chromatin structure and thereby regulating eukaryotic gene transcription. The SET domain proteins represent putative candidates for lysine methyltransferases containing the evolutionarily-conserved SET domain, and important epigenetic regulators present in eukaryotes. In recent years, increasing evidence reveals that SET domain proteins are encoded by a large multigene family in plants and investigation of the SET domain gene family will serve to elucidate the epigenetic mechanism diversity in plants. Although the SET domain gene family has been thoroughly characterized in multiple plant species including two model plant systems, Arabidopsis and rice, through their sequenced genomes, analysis of the entire SET domain gene family in maize was not completed following maize (B73) genome sequencing project. Here, we performed a genome-wide structural and evolutionary analysis of maize SET domain genes from the latest version of the maize (B73) genome. A complete set of 43 SET domain genes (Zmset1-43) were identified in the maize genome using Blast search tools and categorized into seven classes (Class I–VII) based on phylogeny. Chromosomal location of these genes revealed that they are unevenly distributed on all ten chromosomes with seven segmental duplication events, suggesting that segmental duplication played a key role in expansion of the maize SET domain gene family. EST expression data mining revealed that these newly identified genes had temporal and spatial expression pattern and suggested that many maize SET domain genes play functional developmental roles in multiple tissues. Furthermore, the transcripts of the 18 genes (the Class V subfamily) were detected in the leaves by two different abiotic stress treatments using semi-quantitative RT-PCR. The data demonstrated that these genes exhibited different expression levels in stress treatments. Overall, our study will serve to better understand the complexity of the maize SET domain gene family and also be beneficial for future experimental research to further unravel the mechanisms of epigenetic regulation in plants.





Similar content being viewed by others
References
Liu C, Lu F, Cui X, Cao X (2010) Histone methylation in higher plants. Annu Rev Plant Biol 61:395–420
Ng DW, Wang T, Chandrasekharan MB, Aramayo R, Kertbundit S, Hall TC (2007) Plant SET domain-containing proteins: structure, function and regulation. Biochim Biophys Acta 1769:316–329
Kornberg RD, Lorch Y (1999) Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell 98:285–294
Nemeth A, Langst G (2004) Chromatin higher order structure: opening up chromatin for transcription. Brief Funct Genomic Proteomic 2:334–343
Tomschik M, Zheng H, van Holde K, Zlatanova J, Leuba SH (2005) Fast, long-range, reversible conformational fluctuations in nucleosomes revealed by single-pair fluorescence resonance energy transfer. Proc Natl Acad Sci USA 102:3278–3283
Strahl BD, Allis CD (2000) The language of covalent histone modifications. Nature 403:41–45
Zhang Y, Reinberg D (2001) Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev 15:2343–2360
Iizuka M, Smith MM (2003) Functional consequences of histone modifications. Curr Opin Genet Dev 13:154–160
Luger K, Richmond TJ (1998) The histone tails of the nucleosome. Curr Opin Genet Dev 8:140–146
Turner BM (2000) Histone acetylation and an epigenetic code. BioEssays 22:836–845
Jenuwein T, Allis CD (2001) Translating the histone code. Science 293:1074–1080
Chinnusamy V, Zhu JK (2009) Epigenetic regulation of stress responses in plants. Curr Opin Plant Biol 12:133–139
Aquea F, Vega A, Timmermann T, Poupin MJ, Arce-Johnson P (2011) Genome-wide analysis of the SET DOMAIN GROUP family in grapevine. Plant Cell Rep 30:1087–1097
Feng Q, Wang H, Ng HH, Erdjument-Bromage H, Tempst P, Struhl K, Zhang Y (2002) Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr Biol 12:1052–1058
Zhang LS, Ma CR, Ji Q, Wang YF (2009) Genome-wide identification, classification and expression analyses of SET domain gene family in Arabidopsis and rice. Yi Chuan 31:186–198
Pontvianne F, Blevins T, Pikaard CS (2010) Arabidopsis histone lysine methyltransferases. Adv Bot Res 53:1–22
Goodrich J, Puangsomlee P, Martin M, Long D, Meyerowitz EM, Coupland G (1997) A Polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature 386:44–51
Katz A, Oliva M, Mosquna A, Hakim O, Ohad N (2004) FIE and CURLY LEAF polycomb proteins interact in the regulation of homeobox gene expression during sporophyte development. Plant J 37:707–719
Schubert D, Primavesi L, Bishopp A, Roberts G, Doonan J, Jenuwein T, Goodrich J (2006) Silencing by plant Polycomb-group genes requires dispersed trimethylation of histone H3 at lysine 27. EMBO J 25:4638–4649
Baroux C, Gagliardini V, Page DR, Grossniklaus U (2006) Dynamic regulatory interactions of Polycomb group genes: MEDEA autoregulation is required for imprinted gene expression in Arabidopsis. Genes Dev 20:1081–1086
Gehring M, Huh JH, Hsieh TF, Penterman J, Choi Y, Harada JJ, Goldberg RB, Fischer RL (2006) DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell 124:495–506
Jullien PE, Katz A, Oliva M, Ohad N, Berger F (2006) Polycomb group complexes self-regulate imprinting of the Polycomb group gene MEDEA in Arabidopsis. Curr Biol 16:486–492
Chanvivattana Y, Bishopp A, Schubert D, Stock C, Moon YH et al (2004) Interaction of Polycomb-group proteins controlling flowering in Arabidopsis. Development 131:5263–5276
Makarevich G, Leroy O, Akinci U, Schubert D, Clarenz O et al (2006) Different Polycomb group complexes regulate common target genes in Arabidopsis. EMBO Rep 7:947–952
Wang D, Tyson MD, Jackson SS, Yadegari R (2006) Partially redundant functions of two SET-domain polycomb-group proteins in controlling initiation of seed development in Arabidopsis. Proc Natl Acad Sci USA 103:13244–13249
Baumbusch LO, Thorstensen T, Krauss V, Fischer A, Naumann K et al (2001) The Arabidopsis thaliana genome contains at least 29 active genes encoding SET domain proteins that can be assigned to four evolutionarily conserved classes. Nucleic Acids Res 29:4319–4333
Zhao Z, Yu Y, Meyer D, Wu C, Shen WH (2005) Prevention of early flowering by expression of FLOWERING LOCUS C requires methylation of histone H3 K36. Nat Cell Biol 7:1256–1260
Alvarez-Venegas R, Pien S, Sadder M, Witmer X, Grossniklaus U, Avramova Z (2003) ATX-1, an Arabidopsis homolog of trithorax, activates flower homeotic genes. Curr Biol 13:627–637
Alvarez-Venegas R, Avramova Z (2005) Methylation patterns of histone H3 Lys 4, Lys 9 and Lys 27 in transcriptionally active and inactive Arabidopsis genes and in atx1 mutants. Nucleic Acids Res 33:5199–5207
Springer NM, Napoli CA, Selinger DA, Pandey R, Cone KC, Chandler VL, Kaeppler HF, Kaeppler SM (2003) Comparative analysis of SET domain proteins in maize and Arabidopsis reveals multiple duplications preceding the divergence of monocots and dicots. Plant Physiol 132:907–925
Jacob Y, Stroud H, Leblanc C, Feng S, Zhuo L, Caro E, Hassel C, Gutierrez C, Michaels SD, Jacobsen SE (2010) Regulation of heterochromatic DNA replication by histone H3 lysine 27 methyltransferases. Nature 466:987–991
Mutskov V, Felsenfeld G (2004) Silencing of transgene transcription precedes methylation of promoter DNA and histone H3 lysine 9. EMBO J 23:138–149
Huang Yong, Liu Chunlin, Shen Wen-Hui, Ruan Ying (2011) Phylogenetic analysis and classification of the Brassica rapa SET-domain protein family. BMC Plant Biol 11:175
Schnable PS, Ware D, Fulton RS et al (2009) The B73 maize genome:complexity, diversity, and dynamics. Science 326:1112–1115
Finn RD, Mistry J, Schuster-Bockler B, Griffiths-Jones S, Hollich V et al (2006) Pfam: clans, web tools and services. Nucleic Acids Res 34:D247–D251
Letunic I, Doerks T, Bork P (2009) SMART 6: recent updates and new developments. Nucleic Acids Res 37:D229–D232
Thompson J, Higgins D, Gibson T (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5:150–163
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Yang S, Zhang X, Yue J, Tian D, Chen J (2008) Recent duplications dominate NBS-encoding gene expansion in two woody species. Mol Genet Genomics 280:187–198
Gu Z, Cavalcanti A, Chen FC, Bouman P, Li WH (2002) Extent of gene duplication in the genomes of drosophila, nematode, and yeast. Mol Biol Evol 19:256–262
Schultz J, Copley RR, Doerks T, Ponting CP, Bork P (2000) SMART: a Web-based tool for the study of genetically mobile domains. Nucleic Acids Res 28:231–234
Qian YX, Cheng Y, Cheng X, Jiang HY, Zhu SW, Cheng BJ (2011) Identification and characterization of Dicer-like, Argonaute and RNA-dependent RNA polymerase gene families in maize. Plant Cell Rep 30:1347–1363
Aasland R, Gibson TJ, Stewart AF (1995) The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem Sci 20:56–59
Stec I, Nagl SB, van Ommen GJ, den Dunnen JT (2000) The PWWP domain: a potential protein–protein interaction domain in nuclear proteins influencing differentiation? FEBS Lett 473:1–5
Blanc G, Wolfe KH (2004) Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 16:1679–1691
Acknowledgments
This study was supported by grants from the China Postdoctoral Science Foundation (No. 2012M521212) and the Anhui Provincial Natural Science Foundation (No. 1308085MC44) and the Anhui Provincial University Natural Science Research Key Project (No. KJ2013A132) and the Anhui Postdoctoral Science Foundation. We wish to thank the anonymous reviewers for their helpful comments on this manuscript. We also acknowledge Professor Yilong Xi and Professor Beijiu Cheng for critical reading of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Qian, Y., Xi, Y., Cheng, B. et al. Identification and characterization of the SET domain gene family in maize. Mol Biol Rep 41, 1341–1354 (2014). https://doi.org/10.1007/s11033-013-2980-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-013-2980-x