Abstract
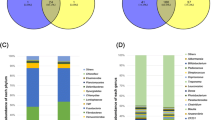
The performance of birds appears to vary among the flock of growing broilers which may in part be due to variation in their gut microbiota. In the view of poultry industry, it is desirable to minimise such variation. We investigated metagenomic profile of fecal bacteria in birds with high and low feed conversion ratio (FCR) to identify microbial community linked to low and high FCR by employing high throughput pyrosequencing of 16S rRNA genomic targets. Therefore feeding trial was investigated in order to identify fecal bacteria consistently linked with better feed conversion ratio in bird performance as measured by body weight gain. High-throughput 16S rRNA gene based pyrosequencing was used to provide a comparative analysis of fecal microbial diversity. The fecal microbial community of birds was predominated by Proteobacteria (48.04 % in high FCR and 49.98 % in low FCR), Firmicutes (26.17 % in high FCR and 36.23 % in low FCR), Bacteroidetes (18.62 % in high FCR and 11.66 % in low FCR), as well as unclassified bacteria (15.77 % in high FCR and 14.29 % in low FCR), suggesting that a large portion of fecal microbiota is novel and could be involved in currently unknown functions. The most prevalent bacterial classes in high FCR and low FCR were Gammaproteobacteria, Clostridia and Bacteroidia. However in low FCR birds Phascolarctobacterium, Faecalibacterium and Clostridium predominated among the Clostridia. In FCR comparison of fecal bacteria, about 36 genera were differentially abundant between high and low FCR birds. This information could be used to formulate effective strategies to improve feed efficiency and feed formulation for optimal gut health.

Similar content being viewed by others
References
Aggrey SE, Karnuah AB, Sebastian B, Anthony NB (2010) Genetic properties of feed efficiency parameters in meat-type chickens. Genet Sel Evol 42:25
Engberg RM, Hedemann MS, Steenfeldt S, Jensen BB (2004) Influence of whole wheat and xylanase on broiler performance and microbial composition and activity in the digestive tract. Poult Sci 83(6):925–938
Havenstein GB, Ferket PR, Qureshi MA (2003) Growth, livability, and feed conversion of 1957 versus 2001 broilers when fed representative 1957 and 2001 broiler diets. Poult Sci 82(10):1500–1508
Pedroso AA, Menten JF, Lambais MR, Racanicci AM, Longo FA, Sorbara JO (2006) Intestinal bacterial community and growth performance of chickens fed diets containing antibiotics. Poult Sci 85(4):747–752
Lu J, Idris U, Harmon B, Hofacre C, Maurer JJ, Lee MD (2003) Diversity and succession of the intestinal bacterial community of the maturing broiler chicken. Appl Environ Microbiol 69(11):6816–6824
Torok VA, Hughes RJ, Ophel-Keller K, Ali M, Macalpine R (2009) Influence of different litter materials on cecal microbiota colonization in broiler chickens. Poult Sci 88(12):2474–2481
Torok VA, Ophel-Keller K, Loo M, Hughes RJ (2008) Application of methods for identifying broiler chicken gut bacterial species linked with increased energy metabolism. Appl Environ Microbiol 74(3):783–791
Lumpkins BS, Batal AB, Lee MD (2010) Evaluation of the bacterial community and intestinal development of different genetic lines of chickens. Poult Sci 89(8):1614–1621
Apajalahti JH, Sarkilahti LK, Maki BR, Heikkinen JP, Nurminen PH, Holben WE (1998) Effective recovery of bacterial DNA and percent-guanine-plus-cytosine-based analysis of community structure in the gastrointestinal tract of broiler chickens. Appl Environ Microbiol 64(10):4084–4088
Hill JE, Seipp RP, Betts M, Hawkins L, Van Kessel AG, Crosby WL, Hemmingsen SM (2002) Extensive profiling of a complex microbial community by high-throughput sequencing. Appl Environ Microbiol 68(6):3055–3066
Hume ME, Kubena LF, Edrington TS, Donskey CJ, Moore RW, Ricke SC, Nisbet DJ (2003) Poultry digestive microflora biodiversity as indicated by denaturing gradient gel electrophoresis. Poult Sci 82(7):1100–1107
Oviedo-Rondon EO, Hume ME, Hernandez C, Clemente-Hernandez S (2006) Intestinal microbial ecology of broilers vaccinated and challenged with mixed Eimeria species, and supplemented with essential oil blends. Poult Sci 85(5):854–860
Parker J, Oviedo-Rondon EO, Clack BA, Clemente-Hernandez S, Osborne J, Remus JC, Kettunen H, Makivuokko H, Pierson EM (2007) Enzymes as feed additive to aid in responses against Eimeria species in coccidia-vaccinated broilers fed corn-soybean meal diets with different protein levels. Poult Sci 86(4):643–653
Apajalahti J, Kettunen A, Graham H (2004) Characteristics of the gastrointestinal microbial communities, with special reference to the chicken. World Poultry Sci J 60(2):223–232
Barnes EM (1979) The intestinal microflora of poultry and game birds during life and after storage. Address of the president of the Society for Applied Bacteriology delivered at a meeting of the society on 10 January 1979. J Appl Bacteriol 46(3):407–419
Bjerrum L, Engberg RM, Leser TD, Jensen BB, Finster K, Pedersen K (2006) Microbial community composition of the ileum and cecum of broiler chickens as revealed by molecular and culture-based techniques. Poult Sci 85(7):1151–1164
Hubener K, Vahjen W, Simon O (2002) Bacterial responses to different dietary cereal types and xylanase supplementation in the intestine of broiler chicken. Arch Tierernahr 56(3):167–187
Torok VA, Allison GE, Percy NJ, Ophel-Keller K, Hughes RJ (2011) Influence of antimicrobial feed additives on broiler commensal posthatch gut microbiota development and performance. Appl Environ Microbiol 77(10):3380–3390
Yin Y, Lei F, Zhu L, Li S, Wu Z, Zhang R, Gao GF, Zhu B, Wang X (2010) Exposure of different bacterial inocula to newborn chicken affects gut microbiota development and ileum gene expression. ISME J 4(3):367–376
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73(16):5261–5526
Coates ME, Fuller R, Harrison GF, Lev M, Suffolk SF (1963) A comparison of the growth of chicks in the Gustafsson germ-free apparatus and in a conventional environment, with and without dietary supplements of penicillin. Br J Nutr 17:141–150
Rebel JM, Van Hemert S, Hoekman AJ, Balk FR, Stockhofe-Zurwieden N, Bakker D, Smits MA (2006) Maternal diet influences gene expression in intestine of offspring in chicken (Gallus gallus). Comp Biochem Physiol A Mol Integr Physiol 145(4):502–508
Brown KH (2003) Diarrhea and malnutrition. J Nutr 133(1):328S–332S
Tiihonen K, Kettunen H, Bento MH, Saarinen M, Lahtinen S, Ouwehand AC, Schulze H, Rautonen N (2010) The effect of feeding essential oils on broiler performance and gut microbiota. Br Poult Sci 51(3):381–392
Clemens J, Albert MJ, Rao M, Qadri F, Huda S, Kay B, van Loon FP, Sack D, Pradhan BA, Sack RB (1995) Impact of infection by Helicobacter pylori on the risk and severity of endemic cholera. J Infect Dis 171(6):1653–1656
Shahinian ML, Passaro DJ, Swerdlow DL, Mintz ED, Rodriguez M, Parsonnel J (2000) Helicobacter pylori and epidemic Vibrio cholerae O1 infection in Peru. Lancet 355(9201):377–378
Benson AK, Kelly SA, Legge R, Ma F, Low SJ, Kim J, Zhang M, Oh PL, Nehrenberg D, Hua K, Kachman SD, Moriyama EN, Walter J, Peterson DA, Pomp D (2010) Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc Nat Acad Sci USA 107(44):18933–18938
Buhnik-Rosenblau K, Danin-Poleg Y, Kashi Y (2011) Predominant effect of host genetics on levels of Lactobacillus johnsonii bacteria in the mouse gut. Appl Environ Microbiol 77(18):6531–6538. doi:10.1128/AEM.00324-11
Gupta SS, Mohammed MH, Ghosh TS, Kanungo S, Nair GB, Mande SS (2011) Metagenome of the gut of a malnourished child. Gut Pathog 3:7
Orcutt B, Bailey B, Staudigel H, Tebo BM, Edwards KJ (2009) An interlaboratory comparison of 16S rRNA gene-based terminal restriction fragment length polymorphism and sequencing methods for assessing microbial diversity of seafloor basalts. Environ Microbiol 11(7):1728–1735
Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI (2006) An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444(7122):1027–1031
Zhang H, DiBaise JK, Zuccolo A, Kudrna D, Braidotti M, Yu Y, Parameswaran P, Crowell MD, Wing R, Rittmann BE, Krajmalnik-Brown R (2009) Human gut microbiota in obesity and after gastric bypass. Proc Natl Acad Sci U S A 106(7):2365–2370
Torok VA, Hughes RJ, Mikkelsen LL, Perez-Maldonado R, Balding K, MacAlpine R, Percy NJ, Ophel-Keller K (2011) Identification and characterization of potential performance-related gut microbiotas in broiler chickens across various feeding trials. Appl Environ Microbiol 77(17):5868–5878
Beckmann L, Simon O, Vahjen W (2006) Isolation and identification of mixed linked beta -glucan degrading bacteria in the intestine of broiler chickens and partial characterization of respective 1,3–1,4-beta -glucanase activities. J Basic Microbiol 46(3):175–185
Collier CT, van der Klis JD, Deplancke B, Anderson DB, Gaskins HR (2003) Effects of tylosin on bacterial mucolysis, Clostridium perfringens colonization, and intestinal barrier function in a chick model of necrotic enteritis. Antimicrob Agents Chemother 47(10):3311–3317
Knarreborg A, Engberg RM, Jensen SK, Jensen BB (2002) Quantitative determination of bile salt hydrolase activity in bacteria isolated from the small intestine of chickens. Appl Environ Microbiol 68(12):6425–6428
Acknowledgments
We thank Department of Biotechnology (DBT), Government of India and Rashtriya Krishi Vikas Yojana (RKVY) Government of Gujarat, for their financial support for this study. Support provided by the Chairman, Marshall Breeders Pvt Limited, Nasik-422001, Maharashtra to conduct the study reported here is also acknowledged with respect and gratitude. Authors wish to thank Dr. A.E. Nivsarkar, Ex-Director, National Bureau of Animal Genetic Resources Karnal for critical reading of the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Singh, K.M., Shah, T., Deshpande, S. et al. High through put 16S rRNA gene-based pyrosequencing analysis of the fecal microbiota of high FCR and low FCR broiler growers. Mol Biol Rep 39, 10595–10602 (2012). https://doi.org/10.1007/s11033-012-1947-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-012-1947-7