Abstract
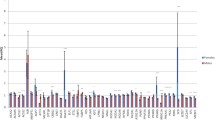
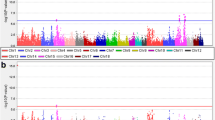
The aim of this research was to screen polymorphism and to perform association study of porcine AMBP (alpha-1-microglobulin/bikunin precursor), GC (group-specific component protein) and PPP1R3B (protein phosphatase 1, regulatory (inhibitor) subunit 3B) genes with meat quality traits as well as to unravel the transcriptional regulation of these genes by expression QTL (eQTL) study. For this purpose, Duroc × Pietrain F2 resource population (DuPi; n = 313) and a commercial breed Pietrain (Pi; n = 110) were used for association and only DuPi for expression and eQTL study. A SNP was identified in the genes AMBP (g.22229C>T), GC (g.398C>T) and PPP1R3B (c.479A>G), respectively. In DuPi SNP of AMBP was associated (P < 0.05) with meat colour, pH1L, pH24L, pH24H and conductivity24L; SNP of GC showed tendency to association (P < 0.10) with pH24H, conductivity1L and thawing loss, and SNP of PPP1R3B was associated (P < 0.05) with meat colour, pH1L, pH24L, pH24H and shear force. In Pi SNPs of AMBP and GC was associated with pH24H and PPP1R3B SNP was associated with pH24L. The mRNA levels in Longissimus dorsi muscle tissue of these three genes were evaluated by using qRT-PCR to identify association between gene expression and meat quality traits as well as to analyse eQTL. The mRNA expression of PPP1R3B associated with pH24L (P < 0.05). Expression of these three genes was higher in animals with low pH of muscle. Linkage analysis using QTL Express revealed ten trans-regulated eQTL on seven porcine autosomes. Suggestive eQTL [P < 0.05, CW (chromosome-wide)] were found for PPP1R3B on SSC3 and 13. These results revealed that genetic variation and gene expression of these genes are associated with the meat quality traits. These three genes could influence meat quality and could be potential positional, physiological and functional candidate gene for meat quality traits in pigs. However, the analysis of eQTL also suggested that we need to consider additional genes encoding for transcription factors (TF), via fine-mapping underlying the eQTL peaks, in order to understand interaction among these genes.


Similar content being viewed by others
References
Ponsuksili S, Jonas E, Murani E, Phatsara C, Srikanchai T, Walz C, Schwerin M, Schellander K, Wimmers K (2008) Trait correlated expression combined with expression qtl analysis reveals biological pathways and candidate genes affecting water holding capacity of muscle. BMC Genomics 9(1):367
Ponsuksili S, Murani E, Phatsara C, Jonas E, Walz C, Schwerin M, Schellander K, Wimmers K (2008) Expression profiling of muscle reveals transcripts differentially expressed in muscle that affect water-holding capacity of pork. J Agric Food Chem 56(21):10311–10317. doi:10.1021/jf800881y
Liu G, Jennen DG, Tholen E, Juengst H, Kleinwachter T, Holker M, Tesfaye D, Un G, Schreinemachers HJ, Murani E, Ponsuksili S, Kim JJ, Schellander K, Wimmers K (2007) A genome scan reveals qtl for growth, fatness, leanness and meat quality in a duroc–pietrain resource population. Anim Genet 38:241–252
Duan YY, Ma JW, Yuan F, Huang LB, Yang KX, Xie JP, Wu GZ, Huang LS (2009) Genome-wide identification of quantitative trait loci for pork temperature, ph decline, and glycolytic potential in a large-scale white duroc x chinese erhualian resource population. J Anim Sci 87(1):9–16. doi:10.2527/jas.2008-1128
Ovilo C, Clop A, Noguera JL, Oliver MA, Barragan C, Rodriguez C, Silio L, Toro MA, Coll A, Folch JM, Sanchez A, Babot D, Varona L, Perez-Enciso M (2002) Quantitative trait locus mapping for meat quality traits in an iberian x landrace f2 pig population. J Anim Sci 80:2801–2808
Velleman SG (2002) Role of the extracellular matrix in muscle growth and development. J Anim Sci 80(E-Suppl_2):E8–E13
Worby CA, Gentry MS, Dixon JE (2008) Malin decreases glycogen accumulation by promoting the degradation of protein targeting to glycogen (ptg). J Biol Chem 283(7):4069–4076. doi:10.1074/jbc.M708712200
Munro S, Cuthbertson DJ, Cunningham J, Sales M, Cohen PT (2002) Human skeletal muscle expresses a glycogen-targeting subunit of pp1 that is identical to the insulin-sensitive glycogen-targeting subunit g(l) of liver. Diabetes 51(3):591–598
Jansen RC, Nap J-P (2001) Genetical genomics: the added value from segregation. Trends Genet 17(7):388–391
Williams JL (2005) The use of marker-assisted selection in animal breeding and biotechnology. Rev Sci Tech Office Int Des Epizoot 24(1):379–391
Srikanchai T, Murani E, Phatsara C, Schwerin M, Schellander K, Ponsuksili S, Wimmers K (2010) Association of zyx polymorphisms with carcass and meat quality traits in commercial pigs meat. Science 84(1):159–164
ZDS (2004) Richtlinie fuer die stationspruefung auf mastleistung, schlachtkoerperwert und fleischbeschaffenheit beim schwein. Zentralverband der Deutschen Schweineproduktion eV, Ausschussfuer Leistungspruefung und Zuchtwertschaetzung, Bonn
Honikel KO (1986) Wasserbindungsvermogen von fleisch. Mitt BAFF 94:7150–7154
Rozen S, Skaletsky H (2000) Primer3 on the www for general users and for biologist programmers. Methods Mol Biol 132:365–386
Große-Brinkhaus C, Phatsara C, Tholen E, Schellander K, Jonas E (2009) Feinkartierung von qtl für fleischqualitätsmerkmale auf dem porcinen chromosom 1. Züchtungskunde 81:63–68
O’Connell JR, Weeks DE (1998) Pedcheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 63(1):259–266
Green P, Falls K, Crooks S (1990) Documentation for cri-map (version 2.4)
Knott SA, Marklund L, Haley CS, Andersson K, Davies W, Ellegren H, Fredholm M, Hansson I, Hoyheim B, Lundstrom K, Moller M, Andersson L (1998) Multiple marker mapping of quantitative trait loci in a cross between outbred wild boar and large white pigs. Genetics 149(2):1069–1080
Seaton G, Haley CS, Knott SA, Kearsey M, Visscher PM (2002) Qtl express: mapping quantitative trait loci in simple and complex pedigrees. Bioinformatics 18(2):339–340
Churchill GA, Doerge RW (1994) Empirical threshold values for quantitative trait mapping. Genetics 138:963–971
Fischer K (2007) Drip loss in pork: influencing factors and relation to further meat quality traits. J Anim Breed Genet 124(Suppl 1):12–18. doi:10.1111/j.1439-0388.2007.00682.x
Lopez-Bote C, Warriss PD (1988) A note on the relationships between measures of water holding capacity in the m. Longissimus dorsi and total drip loss from butchered pig carcasses during storage meat. Science 23(3):227–234
Byrne CE, Troy DJ, Buckley DJ (2000) Postmortem changes in muscle electrical properties of bovine m. longissimus dorsi and their relationship to meat quality attributes and ph fall. Meat Sci 53(1):23–34
Tyagi S, Salier J-P, Lal SK (2002) The liver-specific human alpha1-microglobulin/bikunin precursor (ambp) is capable of self-association. Arch Biochem Biophys 399(7):66–72
Grewal JS, Tsai JY, Khan SR (2005) Oxalate-inducible ambp gene and its regulatory mechanism in renal tubular epithelial cells. Biochem J 387(3):609–616
Velleman SG (2000) The role of the extracellular matrix in skeletal development. Poult Sci 79(7):985–989
Otterbein LR, Cosio C, Graceffa P, Dominguez R (2002) Crystal structures of the vitamin d-binding protein and its complex with actin: structural basis of the actin-scavenger system. Proc Natl Acad Sci USA 99(12):8003–8008. doi:10.1073/pnas.122126299
Huff-Lonergan E, Lonergan SM (2007) Drip loss and water holding capacity of porcine meat. J Anim Breed Genet 124(s1):19–26
Goldschmidt-Clermont PJ, Van Baelen H, Bouillon R, Shook TE, Williams MH, Nel AE, Galbraith RM (1988) Role of group-specific component (vitamin d binding protein) in clearance of actin from the circulation in the rabbit. J Clin Invest 81(5):1519–1527
Foote MR, Horst RL, Huff-Lonergan EJ, Trenkle AH, Parrish FC Jr, Beitz DC (2004) The use of vitamin d3 and its metabolites to improve beef tenderness. J Anim Sci 82(1):242–249
Sandercock DA, Barker ZE, Mitchell MA, Hocking PM (2009) Changes in muscle cell cation regulation and meat quality traits are associated with genetic selection for high body weight and meat yield in broiler chickens. Genet Sel Evol 41:8. doi:10.1186/1297-9686-41-8
Thomsen H, Lee HK, Rothschild MF, Malek M, Dekkers JC (2004) Characterization of quantitative trait loci for growth and meat quality in a cross between commercial breeds of swine. J Anim Sci 82(8):2213–2228
Wimmers K, Fiedler I, Hardge T, Murani E, Schellander K, Ponsuksili S (2006) Qtl for microstructural and biophysical muscle properties and body composition in pigs. BMC Genet 7(1):15
Doherty MJ, Moorhead G, Morrice N, Cohen P, Cohen PT (1995) Amino acid sequence and expression of the hepatic glycogen-binding (gl)-subunit of protein phosphatase-1. FEBS Lett 375(3):294–298
Choe JH, Choi YM, Lee SH, Shin HG, Ryu YC, Hong KC, Kim BC (2008) The relation between glycogen, lactate content and muscle fiber type composition, and their influence on postmortem glycolytic rate and pork quality. Meat Sci 80(2):355–362
Edwards DB, Bates RO, Osburn WN (2003) Evaluation of duroc- vs. pietrain-sired pigs for carcass and meat quality measures. J Anim Sci 81(8):1895–1899
Jennen DG, Brings AD, Liu G, Jungst H, Tholen E, Jonas E, Tesfaye D, Schellander K, Phatsara C (2007) Genetic aspects concerning drip loss and water-holding capacity of porcine meat. J Anim Breed Genet 124(Suppl 1):2–11. doi:10.1111/j.1439-0388.2007.00681.x
Cooke NE, McLeod JF, Wang XK, Ray K (1991) Vitamin d binding protein: genomic structure, functional domains, and mRNA expression in tissues. J Steroid Biochem Mol Biol 40(4–6):787–793
Montori-Grau M, Guitart M, Lerin C, Andreu AL, Newgard CB, Garcia-Martinez C, Gomez-Foix AM (2007) Expression and glycogenic effect of glycogen-targeting protein phosphatase 1 regulatory subunit gl in cultured human muscle. Biochem J 405(1):107–113. doi:10.1042/BJ20061572
Yoshidaa K, Suzukib Y, Yamamotoa K, Sinohara H (1999) Guinea pig α1-microglobulin/bikunin: Cdna sequencing, tissue expression and expression during acute phase. Comp Biochem Physiol Part B 122(2):165–172
Kwasiborski A, Rocha D, Terlouw C (2009) Gene expression in large white or duroc-sired female and castrated male pigs and relationships with pork quality. Anim Genet 40(6):852–862. doi:10.1111/j.1365-2052.2009.01925.x
Wang YH, Byrne KA, Reverter A, Harper GS, Taniguchi M, McWilliam SM, Mannen H, Oyama K, Lehnert SA (2005) Transcriptional profiling of skeletal muscle tissue from two breeds of cattle. Mamm Genome 16(3):201–210. doi:10.1007/s00335-004-2419-8
Xu X, Qiu H, Du ZQ, Fan B, Rothschild MF, Yuan F, Liu B (2010) Porcine csrp3: polymorphism and association analyses with meat quality traits and comparative analyses with csrp1 and csrp2. Mol Biol Rep 37(1):451–459. doi:10.1007/s11033-009-9632-1
Guignota F, Touraillea C, Oualia A, Renerrea M, Monin G (1994) Relationships between post-mortem pH changes and some traits of sensory quality in veal. Meat Sci 37(3):315–325
Gerbens F, Verburg FJ, Van Moerkerk HT, Engel B, Buist W, Veerkamp JH, te Pas MF (2001) Associations of heart and adipocyte fatty acid-binding protein gene expression with intramuscular fat content in pigs. J Anim Sci 79(2):347–354
Michaelson JJ, Loguercio S, Beyer A (2009) Detection and interpretation of expression quantitative trait loci (eqtl). Methods 48(3):265–276. doi:10.1016/j.ymeth.2009.03.004
Hubner N, Wallace CA, Zimdahl H, Petretto E, Schulz H, Maciver F, Mueller M, Hummel O, Monti J, Zidek V, Musilova A, Kren V, Causton H, Game L, Born G, Schmidt S, Muller A, Cook SA, Kurtz TW, Whittaker J, Pravenec M, Aitman TJ (2005) Integrated transcriptional profiling and linkage analysis for identification of genes underlying disease. Nat Genet 37:243–253
Morley M, Molony CM, Weber TM, Devlin JL, Ewens KG, Spielman RS, Cheung VG (2004) Genetic analysis of genome-wide variation in human gene expression. Nature 430(7001):743–747
Ponsuksili S, Murani E, Phatsara C, Schwerin M, Schellander K, Wimmers K (2010) Expression quantitative trait loci analysis of genes in porcine muscle by quantitative real-time rt-pcr compared to microarray data. Heredity 105(3):309–317. doi:10.1038/hdy.2010.5
Stranger BE, Forrest MS, Clark AG, Minichiello MJ, Deutsch S, Lyle R, Hunt S, Kahl B, Antonarakis SE, Tavare S, Deloukas P, Dermitzakis ET (2005) Genome-wide associations of gene expression variation in humans. PLoS Genet 1(6):e78. doi:10.1371/journal.pgen.0010078
Bystrykh L, Weersing E, Dontje B, Sutton S, Pletcher MT, Wiltshire T, Su AI, Vellenga E, Wang J, Manly KF, Lu L, Chesler EJ, Alberts R, Jansen RC, Williams RW, Cooke MP, de Haan G (2005) Uncovering regulatory pathways that affect hematopoietic stem cell function using ‘genetical genomics’. Nat Genet 37(3):225–232. doi:10.1038/ng1497
Gibson G, Weir B (2005) The quantitative genetics of transcription. Trends Genet 21(11):616–623. doi:10.1016/j.tig.2005.08.010
Petretto E, Mangion J, Dickens NJ, Cook SA, Kumaran MK, Lu H, Fischer J, Maatz H, Kren V, Pravenec M, Hubner N, Aitman TJ (2006) Heritability and tissue specificity of expression quantitative trait loci. PLoS Genet 2(10):e172. doi:10.1371/journal.pgen.0020172
Giangrande PH, Zhu W, Schlisio S, Sun X, Mori S, Gaubatz S, Nevins JR (2004) A role for e2f6 in distinguishing g1/s- and g2/m-specific transcription. Genes Dev 18(23):2941–2951. doi:10.1101/gad.1239304
Wang X, Blagden C, Fan J, Nowak SJ, Taniuchi I, Littman DR, Burden SJ (2005) Runx1 prevents wasting, myofibrillar disorganization, and autophagy of skeletal muscle. Genes Dev 19(14):1715–1722. doi:10.1101/gad.1318305
Ponsuksili S, Chomdej S, Murani E, Blaser U, Schreinemachers HJ, Schellander K, Wimmers K (2005) Snp detection and genetic mapping of porcine genes encoding enzymes in hepatic metabolic pathways and evaluation of linkage with carcass traits. Anim Genet 36(6):477–483. doi:10.1111/j.1365-2052.2005.01351.x
Malek M, Dekkers JC, Lee HK, Baas TJ, Prusa K, Huff-Lonergan E, Rothschild MF (2001) A molecular genome scan analysis to identify chromosomal regions influencing economic traits in the pig. Ii. Meat and muscle composition. Mamm Genome 12:637–645
Lee SS, Chen Y, Moran C, Cepica S, Reiner G, Bartenschlager H, Moser G, Geldermann H (2003) Linkage and qtl mapping for sus scrofa chromosome 2. J Anim Breed Genet 120:11–19
Heuven HC, van Wijk RH, Dibbits B, van Kampen TA, Knol EF, Bovenhuis H (2009) Mapping carcass and meat quality qtl on sus scrofa chromosome 2 in commercial finishing pigs. Genet Sel Evol 41:4. doi:10.1186/1297-9686-41-4
Edwards DB, Ernst CW, Raney NE, Doumit ME, Hoge MD, Bates RO (2008) Quantitative trait locus mapping in an f2 duroc × pietrain resource population: Ii. Carcass and meat quality traits. J Anim Sci 86(2):254–266. doi:10.2527/jas.2006-626
van Wijk HJ, Dibbits B, Baron EE, Brings AD, Harlizius B, Groenen MA, Knol EF, Bovenhuis H (2006) Identification of quantitative trait loci for carcass composition and pork quality traits in a commercial finishing cross. J Anim Sci 84:789–799
Acknowledgments
This project was supported by the project, “Functional Genetic Principles of Water Binding Capacity in Pork (DRIP)” under key project DFG-Forschergruppe “DRIP” FOR 753, Germany. The authors are indebted to the German Research Foundation. The authors are thankful to Nadine Leyer, Institute of Animal Science, University of Bonn, Germany for her technical assistance during experiment.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Cinar, M.U., Kayan, A., Uddin, M.J. et al. Association and expression quantitative trait loci (eQTL) analysis of porcine AMBP, GC and PPP1R3B genes with meat quality traits. Mol Biol Rep 39, 4809–4821 (2012). https://doi.org/10.1007/s11033-011-1274-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-011-1274-4