Abstract
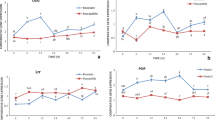
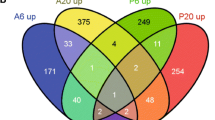
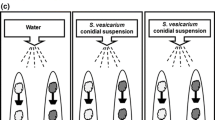
The initial phases of the disease establishment are very crucial for the compatible interactions. Pathogens must overcome the responses generated by the host for the onset of disease invasion. The compatible interaction is inadequately represented in plant-pathogen interaction studies. To gain broader insight into the early responses elicited by chickpea blight fungus Ascochyta rabiei during compatible interaction; we isolated early responsive genes of chickpea using PCR based suppression subtractive hybridization (SSH) strategy. We obtained ~250 unique genes after homology search and redundancy elimination. Based on their potential cellular functions, these genes were broadly classified into eleven different categories viz. stress, signaling, gene regulation, cellular metabolism and genes of unknown functions. Present study revealed few unexpected genes which have a possible role in induced immunity and disease progression. We employed macroarray, northern blot, real-time PCR and cluster analysis to develop transcript profiles. Most of the genes analyzed were early induced and were transcriptionally upregulated upon 24 h post inoculation. Our approach has rendered the isolation of early responsive genes involved in signaling and regulation of metabolic changes upon fungal infection. The information obtained will help to dissect the molecular mechanisms during compatible chickpea–Ascochyta interactions.



Similar content being viewed by others
References
Feng H, Wang X, Sun Y, Wang X, Chen X, Guo J, Duan Y, Huang L, Kang Z (2011) Cloning and characterization of a calcium binding EF-hand protein gene TaCab1 from wheat and its expression in response to Puccinia striiformis f. sp. tritici and abiotic stresses. Mol Biol Rep 38:3857–3866
Oliver RP, Ipcho SVS (2004) Arabidopsis pathology breathes new life into the necrotrophs-vs. biotrophs classification of fungal pathogens. Mol Plant Pathol 5:347–352
Markham JE, Hille J (2001) Host selective toxins as agents of cell death in plant–fungus interactions. Mol Plant Pathol 2:229–239
Wolpert TJ, Dunkle LD, Ciuffetti LM (2002) Host-selective toxins and avirulence determinants: what’s in a name? Annu Rev Phytopathol 40:251–285
Staples RC, Mayer AM (2003) Suppression of host resistance by fungal plant pathogens. Isr J Plant Sci 51:175–186
Govrin EM, Levine A (2000) The hypersensitive response facilitates plant infection by the necrotrophic pathogen Botrytis cinerea. Curr Biol 10:751–757
Jones JDG, Dangl JL (2006) The plant immune system. Nature 444:323–329
Chisholm ST, Coaker G, Day B, Staskawicz BJ (2006) Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124:803–814
Faris JD, Zhang Z, Lu H, Reddy L, Cloutier S, Fellers JP, Meinhardt SW, Rasmussen JB, Xu SS, Oliver RP, Simons KJ, Friesen TL (2010) A unique wheat disease resistance-like gene governs effector triggered susceptibility to necrotrophic pathogens. Proc Natl Acad Sci USA 107:13544–13549
Panstruga R (2003) Establishing compatibility between plants and obligate biotrophic pathogens. Curr Opin Plant Biol 6:320–326
Nene YL, Reddy MV (1987) Chickpea diseases and their control. In: Saxena MC, Singh KB (eds) The chickpea. CAB International, UK, pp 233–270
Gaur RB, Singh RD (1996) Effects of Ascochyta blight on grain yield and protein in chickpea. Indian J Mycol Plant Pathol 26:259–262
Jayakumar P, Gossen BD, Gan YT, Warkentin TD, Banniza S (2005) Ascochyta blight of chickpea: infection and host resistance mechanisms. Can J Plant Pathol 27:499–509
Armstromg-Cho C, Gossen BD (2005) Impact of glandular hair exudates on infection of chickpea by Ascochyta rabiei. Can J Bot 83:22–27
Daniel S, Tiemann K, Wittkampf U, Bless W, Hinderer W, Barz W (1990) Elicitor-induced metabolic changes in cell cultures of chickpea (Cicer arietinum L.) cultivars resistant and susceptible to Ascochyta rabiei: I. Investigations of enzyme activities involved in isoflavone and pterocarpan phytoalexin biosynthesis. Planta 182:270–278
Kessmann H, Barz W (1987) Accumulation of isoflavone and pterocarpan phytoalexins in cell suspension cultures of different cultivars of chickpea. Plant Cell Rep 6:55–59
Khirbat SK, Jalali BL (1998) Production of phytoalexin in the leaves of chickpea (Cicer arietinum L.) after inoculation with Ascochyta rabiei. Legume Res 21:135–143
Cobos MJ, Rubio J, Strange RN, Moreno MT, Gil J, Millan T (2006) A new QTL for ascochyta blight resistance in an RIL population derived from an interspecific cross in chickpea. Euphytica 149:105–111
Flandez-Galvez H, Ades PK, Ford R, Pang ECK, Taylor PWJ (2003) QTL analysis for ascochyta blight resistance in an intraspecific population of chickpea (Cicer arietinum L.). Theor Appl Genet 107:1257–1265
Iruela M, Rubio J, Barro F, Cubero JI, Millan T, Gil J (2006) Detection of two quantitative trait loci for resistance to ascochyta blight in an intra-specific cross of chickpea (Cicer arietinum L.): development of SCAR markers associated with resistance. Theor Appl Genet 112:278–287
Santra DK, Tekeoglu M, Ratnaparkhe M, Kaiser WJ, Muehlbauer FJ (2000) Identification and mapping of QTLs conferring resistance to ascochyta blight in chickpea. Crop Sci 40:1606–1612
Tekeoglu M, Isik M, Muehlbauer FJ (2004) QTL analysis of ascochyta blight resistance in chickpea. Turk J Agric For 28:183–187
Mantri NL, Ford R, Coram TE, Pang ECK (2007) Transcriptional profiling of chickpea genes differentially regulated in response to high-salinity, cold and drought. BMC Genomics 8:303
Singh A, Singh IK, Verma PK (2008) Differential transcript accumulation in Cicer arietinum L. in response to a chewing insect Helicoverpa armigera and defence regulators correlate with reduced insect performance. J Exp Bot 59:2379–2392
Ashraf N, Ghai D, Barman P, Basu S, Gangisetty N, Mandal MK, Chakraborty N, Datta A, Chakraborty S (2009) Comparative analyses of genotype dependent expressed sequence tags and stress-responsive transcriptome of chickpea wilt illustrate predicted and unexpected genes and novel regulators of plant immunity. BMC Genomics 10:415
Huang JY, Jie ZJ, Wang LJ, Yan XH, Wei WH (2011) Analysis of the differential expression of the genes related to Brassica napus seed development. Mol Biol Rep 38:1055–1061
Sun P, Guo Y, Qi J, Zhou L, Li X (2010) Isolation and expression analysis of tuberous root development related genes in Rehmannia glutinosa. Mol Biol Rep 37:1069–1079
Gu L, Xu D, You T, Li X, Yao S, Chen S, Zhao J, Lan H, Zhang F (2011) Analysis of gene expression by ESTs from suppression subtractive hybridization library in Chenopodium album L. under salt stress. Mol Biol Rep. doi:10.1007/s11033-011-0678-5
Sharma K, Mishra AK, Misra RS (2009) Identification and characterization of differentially expressed genes in the resistance reaction in taro infected with Phytophthora colocasiae. Mol Biol Rep 36:1291–1297
Coram TE, Pang ECK (2006) Expression profiling of chickpea genes differentially regulated during a resistance response to Ascochyta rabiei. Plant Biotechnol J 4:647–666
Cornels H, Ichinose Y, Barz W (2000) Characterization of cDNAs encoding two glycine-rich proteins in chickpea (Cicer arietinum L.): accumulation in response to fungal infection and other stress factors. Plant Sci 154:83–88
Ichinose YK, Toyoda BarzW (1999) cDNA cloning and gene expression of three small GTP-binding proteins in defense response of chickpea. Biochim Biophys Acta 1489:462–466
Hanselle T, Ichinose Y, Barz W (2001) Biochemical and molecular biological studies on infection (Ascochyta rabiei)-induced thaumatin-like proteins from chickpea plants (Cicer arietinum L.). Z Naturforsch 56:1095–1107
Rea G, Metoui O, Infantino A, Federico R, Angelini R (2002) Copper amine oxidase expression in defense responses to wounding and Ascochyta rabiei invasion. Plant Physiol 128:865–875
Boominathan P, Shukla R, Kumar A, Manna D, Negi D, Verma PK, Chattopadhyay D (2004) Long term transcript accumulation during the development of dehydration adaptation in Cicer arietinum. Plant Physiol 135:1608–1620
Sambrook J, Russell DW (2001) Molecular cloning-a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Diatchenko L, Lau YF, Campbell AP, Chenchik A, Moqadam F, Huang B, Lukyanov S, Lukyanov K, Gurskaya N, Sverdlov ED, Siebert PD (1996) Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc Natl Acad Sci USA 93:6025–6030
Shi J-l, Wang Y-j, Zhu Z-g, Zhang C-h (2010) The EST Analysis of a suppressive subtraction cDNA library of chinese wild Vitis pseudoreticulata inoculated with Uncinula necator. Agric Sci China 9:233–241
Fossdal CG, Nagy NE, Johnsen O, Dalen LS (2007) Local and systemic stress responses in Norway spruce: similarities in gene expression between a compatible pathogen interaction and drought stress. Physiol Mol Plant Pathol 70:161–173
Thomma BPHJ, Eggermont K, Penninckx IAMA, Mauch-Mani B, Vogelsang R, Cammue BPA, Broekaert WF (1998) Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc Natl Acad Sci USA 95:15107–15111
Xie D, Feys BF, James S, Nieto-Rostro M, Turner JG (1998) COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280:1091–1094
Yan J, Zhang C, Gu M, Bai Z, Zhang W, Qi T, Cheng Z, Peng W, Luo H, Nan F, Wang Z, Xie D (2009) The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. Plant Cell 21:2220–2236
Devoto A, Nieto-Rostro M, Xie D, Ellis C, Harmston R, Patrick E, Davis J, Sherratt L, Coleman M, Turner JG (2002) COI1 links jasmonate signalling and fertility to the SCF ubiquitin-ligase complex in Arabidopsis. Plant J 32:457–466
Steele CL, Gijzen M, Qutob D, Dixon RA (1999) Molecular characterization of the enzyme catalyzing the aryl migration reaction of isoflavonoid biosynthesis in soybean. Arch Biochem Biophys 367:146–150
Aoki T, Akashi T, Ayabe S (2000) Flavonoids of leguminous plants: structure, biological activity, and biosynthesis. J Plant Res 113:475–488
Cheng NH, Hirschi KD (2003) Cloning and characterization of CXIP1, a novel PICOT domain containing Arabidopsis protein that associates with CAX1. J Biol Chem 278:6503–6509
Munns R (2005) Genes and salt tolerance: bringing them together. New Phytol 167:645–663
Fry SC, Smith RC, Renwick KF, Martin DJ, Hodge SK, Matthews KJ (1992) Xyloglucan endotransglycosylase, a new wall-loosening enzyme activity from plants. Biochem J 282:821–828
Eklof JM, Brumer H (2010) The XTH gene family: an update on enzyme structure, function, and phylogeny in xyloglucan remodeling. Plant Physiol 153:456–466
Hunter T (1995) Protein kinases and phosphatases: the yin and yang of protein phosphorylation and signaling. Cell 80:225–236
Pandey SP, Somssich IE (2009) The role of WRKY transcription factors in plant immunity. Plant Physiol 150:1648–1655
Shimada Y, Wu GJ, Watanabe A (1998) A protein encoded by din1, a dark-inducible and senescence-associated gene of radish, can be imported by isolated chloroplasts and has sequence similarity to sulfide dehydrogenase and other small stress proteins. Plant Cell Physiol 39:139–143
He YH, Gan SS (2002) A gene encoding an acyl hydrolase is involved in leaf senescence in Arabidopsis. Plant Cell 14:805–815
Quirino BF, Normanly J, Amasino RM (1999) Diverse range of gene activity during Arabidopsis thaliana leaf senescence includes pathogen-independent induction of defense-related genes. Plant Mol Biol 40:267–278
Buchanan-Wollaston V, Ainsworth C (1997) Leaf senescence in Brassica napus: cloning of senescence related genes by subtractive hybridization. Plant Mol Biol 33:821–834
Quirino BF, Noh YS, Himelblau E, Amasino RM (2000) Molecular aspects of leaf senescence. Trends Plant Sci 5:278–282
Jiang H, Song W, Li A, Yang X, Sun D (2011) Identification of genes differentially expressed in cauliflower associated with resistance to Xanthomonas campestris pv. Campestris. Mol Biol Rep 38:621–629
Kufryk G, Hernandez-Prieto MA, Kieselbach T, Miranda H, Vermaas W, Funk C (2008) Association of small CAB-like proteins (SCPs) of Synechocystis sp. PCC 6803 with Photosystem II. Photosynth Res 95:135–145
Hecht V, Knowles CL, Schoor JKV, Liew LC, Jones SE, Lambert MJM, Weller JL (2007) Pea LATE BLOOMER1 is a GIGANTEA ortholog with roles in photoperiodic flowering, deetiolation, and transcriptional regulation of circadian clock gene homologs. Plant Physiol 144:648–661
Mahalingam R, Gomez-Buitrago A, Eckardt N, Shah N, Guevara-Garcia A, Day P, Raina R, Fedoroff NV (2003) Characterizing the stress/defense transcriptome of Arabidopsis. Genome Biol 4:R20
Jantasuriyarat C, Gowda M, Haller K, Hatfield J, Lu G, Stahlberg E, Zhou B, Li H, Kim H, Yu Y, Dean RA, Wing RA, Soderlund C, Wang GL (2005) Large-scale identification of expressed sequence tags involved in rice and rice blast fungus interaction. Plant Physiol 138:105–115
Acknowledgments
This work was supported by the research grant provided by Department of Biotechnology, Government of India and National Institute of Plant Genome Research, New Delhi. Authors acknowledge Dr. Birendra Singh (Indian Agricultural Research Institute) and Mr. Ashok Kumar (National Institute of Plant Genome Research) for technical help.
Author information
Authors and Affiliations
Corresponding author
Additional information
Jyothi Reddy Cheruku, Kamal Kumar, Saurabh Yadav and Archana Singh have equally contributed in this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
11033_2011_1255_MOESM2_ESM.doc
Table S2 Functional annotation of the unigenes from SSH library. The cDNA sequences of all unigenes obtained in 24 h and 3 h SSH libraries were submitted to the GenBank database and the assigned Accession no. was mentioned. BlastX searches were conducted to determine homologous genes and the putative function. BlastN was performed for cDNAs which did not show any significant homology in BlastX. GenBank Accession no. marked with (*) are cDNAs that showed significant homology in BlastN (e-value cutoff ≥ 1e-10) instead of BlastX (e-value cutoff ≥ 1e-5) (DOC 265 kb)
Rights and permissions
About this article
Cite this article
Jaiswal, P., Cheruku, J.R., Kumar, K. et al. Differential transcript accumulation in chickpea during early phases of compatible interaction with a necrotrophic fungus Ascochyta rabiei . Mol Biol Rep 39, 4635–4646 (2012). https://doi.org/10.1007/s11033-011-1255-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-011-1255-7