Abstract
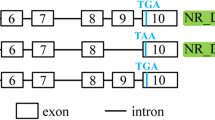
The signaling lymphocyte-activating molecule family 7 (SLAMF7) proteins serve as adhesion molecules on the surface of a variety of mature hematopoietic cells, and also partially control certain innate and adaptive immune responses. We characterized three novel bovine SLAMF7 splice variants, designated as SLAMF7-AS1, AS2, and AS3. All three novel SLAMF7 isoforms are derived from the complete transcripts (SLAMF7-complete) via alternative splicing (AS). The patterns of the three splice variants are exon skipping and alternative 5′ splice sites. Bovine SLAMF7 transcripts are expressed in mammary tissue, as demonstrated by real-time PCR. The levels of the complete transcript expression in the normal mammary tissues were higher than that in Staphylococcus aureus (Staph. aureus)-induced mastitis mammary tissues. However, it was not significant for the mRNA expression level comparison between these two kinds of mammary. The SLAMF-AS2 isoforms are expressed the lowest levels among the three transcripts in both normal and infected mammary tissues. This study provides clues for a better understanding of bovine SLAMF7 gene function.



Similar content being viewed by others
References
Bouchon A, Cella M, Grierson HL, Cohen JI, Colonna M (2001) Cutting edge: activation of NK cell-mediated cytotoxicity by a SAP-independent receptor of the CD2 family. J Immunol 167:5517–5521
Veillette A, Latour S (2003) The SLAM family of immune-cell receptors. Curr Opin Immunol 15:277–285
Stark S, Watzl C (2006) 2B4 (CD244), NTB-A and CRACC (CS1) stimulate cytotoxicity but no proliferation in human NK cells. Int Immunol 18:241–247
Lee JK, Mathew SO, Vaidya SV, Kumaresan PR, Mathew PA (2007) CS1 (CRACC, CD319) induces proliferation and autocrine cytokine expression on human B lymphocytes. J Immunol 179:4672–4678
Calpe S, Wang N, Romero X, Berger SB, Lanyi A, Engel P, Terhorst C (2008) The SLAM and SAP gene families control innate and adaptive immune responses. Adv Immunol 97:177–250
Zimin AV, Delcher AL, Florea L, Kelley DR, Schatz MC, Puiu D, Hanrahan F, Pertea G, Van Tassell CP, Sonstegard TS, Marçais G, Roberts M, Subramanian P, Yorke JA, Salzberg SL (2009) A whole-genome assembly of the domestic cow, Bos taurus. Genome Biol 10(4):R42
Tovar V, del Valle J, Zapater N, Martin M, Romero X, Pizcueta P, Bosch J, Terhorst C, Engel P (2002) Mouse novel Ly9: a new member of the expanding CD150 (SLAM) family of leukocyte cell-surface receptors. Immunogenetics 54:394–402
Sammeth M, Foissac S, Guigo R (2008) A general definition and nomenclature for alternative splicing events. PLoS Comput Biol 4(8):e1000147
Garcia-Blanco MA, Baraniak AP, Lasda EL (2004) Alternative splicing in disease and therapy. Nat Biotechnol 22:535–546
Ast G (2004) How did alternative splicing evolve? Nat Rev Genet 5(10):773–782
Chacko E, Ranganathan S (2009) Genome-wide analysis of alternative splicing in cow: implications in bovine as a model for human diseases. BMC Genom 10(Suppl 3):S11
Keren H, Lev-Maor G, Ast G (2010) Alternative splicing and evolution: diversification, exon definition and function. Nat Rev Genet 11(5):345–355
Jain NC (1979) Common mammary pathogens and factors in infection and mastitis. J Dairy Sci 62:128–134
Swanson KM, Stelwagen K, Dobson J, Henderson HV, Davis SR, Farr VC, Singh K (2009) Transcriptome profiling of Streptococcus uberis-induced mastitis reveals fundamental differences between immune gene expression in the mammary gland and in a primary cell culture model. J Dairy Sci 92(1):117–129
Arnold K, Bordoli L, Kopp J, Schwede T (2006) The SWISS-MODEL Workspace: a web-based environment for protein structure homology modeling. Bioinformatics 22:195–201
Larionov A, Krauseand A, Miller W (2005) A standard curve based method for relative real time PCR data processing. BMC Bioinform 6:62–78
Graveley BR (2001) Alternative splicing: increasing diversity in the proteomic world. Trends Genet 17(2):100–107
Swami M (2009) Alternative splicing: deciding between the alternatives. Nat Rev Genet 10:71
Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB (2008) Alternative isoform regulation in human tissue transcriptomes. Nature 456:470–476
Lee JK, Boles KS, Mathew PA (2004) Molecular and functional characterization of a CS1 (CRACC) splice variant expressed in human NK cells that does not contain immunoreceptor tyrosine-based switch motifs. Eur J Immunol 34:2791–2799
Boles KS, Mathew PA (2001) Molecular cloning of CS1, a novel human natural killer cell receptor belonging to the CD2 subset of the immunoglobulin superfamily. Immunogenetics 52:302–307
Rinaldi M, Li RW, Capuco AV (2010) Mastitis associated transcriptomic disruptions in cattle. Vet Immunol Immunopathol 138(4):267–279
Wellenberg GJ, van der Poel WH, Van Oirschot JT (2002) Viral infections and bovine mastitis: a review. Vet Microbiol 88(1):27–45
Zheng J, Anjanette DW, David EK (2006) Genome-wide expression analysis of lipopolysaccharide-induced mastitis in a mouse model. Infect Immun 74:1907–1915
Caceres JF, Kornblihtt AR (2002) Alternative splicing: multiple control mechanisms and involvement in human disease. Trends Genet 18(4):186–193
Musunuru K (2003) Cell-specific RNA-binding proteins in human disease. Trends Cardiovasc Med 13(5):188–195
Faustino NA, Cooper TA (2003) Pre-mRNA splicing and human disease. Genes Dev 17:419–437
Yan G, Fukabori Y, McBride G, Nikolaropolous S, McKeehan WL (1993) Exon switching and activation of stromal and embryonic fibroblast growth factor (FGF)-FGF receptor genes in prostate epithelial cells accompany stromal independence and malignancy. Mol Cell Biol 13:4513–4522
Acknowledgments
This research was supported by Key Projects in the National Science and Technology Pillar Program during the Twelfth Five-year Plan Period (2011BAD19B04), National Natural Science Foundation (No. 31000543), Modern Agro-industry Technology Research System (No. CARS-37), Well-bred Project from Shandong Province (No. 2009LZ015), and Key Scientific and Technological Project from Shandong province (No. 2009GG20002033).
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ju, Z., Wang, C., Li, Q. et al. Alternative splicing and mRNA expression analysis of bovine SLAMF7 gene in healthy and mastitis mammary tissues. Mol Biol Rep 39, 4155–4161 (2012). https://doi.org/10.1007/s11033-011-1198-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-011-1198-z