Abstract
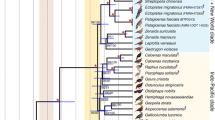
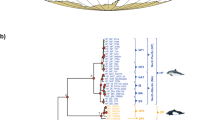
Galloanserae is an ancient and diverse avian group, for which comprehensive molecular evidence relevant to phylogenetic analysis in the context of molecular chronology is lacking. In this study, we present two additional mitochondrial genome sequences of Galloanserae (the whistling duck, Dendrocygna javanica, and the black swan, Cygnus atratus) to broaden the scope of molecular phylogenetic reconstruction. The lengths of the whistling duck’s and black swan’s mitochondrial genomes are 16,753 and 16,748 bases, respectively. Phylogenetic analyses suggest that Dendrocygna is more likely to be in a basal position of the branch consisting of Anatinae and Anserinae, an affiliation that does not conform to its traditional classification. Bayesian approaches were employed to provide a rough timescale for Galloanserae evolution. In general, a narrow range of 95% confidence intervals gave younger estimates than those based on limited genes and estimated that at least two lineages originated before the Coniacian epoch around 90 MYA, well before the Cretaceous-Tertiary boundary. In addition, these results, which were compatible with estimates from fossil evidence, also imply that the origin of numerous genera in Anseriformes took place in the late Oligocene to early Miocene. Taken together, the results presented here provide a working framework for future research on Galloanserae evolution, and they underline the utility of whole mitochondrial genome sequences for the resolution of deep divergence.



Similar content being viewed by others
Abbreviations
- ATP6, and 8:
-
ATPase subunits 6 and 8
- bp:
-
Base pair(s)
- COI-III:
-
Cytochrome c oxidase subunits I–III
- COB :
-
Cytochrome b
- mtDNA:
-
Mitochondrial DNA
- ND1-6, and 4L:
-
NADH dehydrogenase subunits 1–6 and 4L
- rRNA:
-
Ribosomal RNA
- tRNA:
-
Transfer RNA
- LB:
-
Luria–Bertani (medium)
- K/T:
-
Cretaceous-Tertiary
- MYA:
-
Million years ago
- RSCUs:
-
Values of relative synonymous codon usage
- IPTG:
-
Isopropyl β-d-thiogalactopyranoside
References
Boore JL (1999) Animal mitochondrial genomes. Nucleic Acids Res 27:1767–1780
Curole JP, Kocher TD (1999) Mitogenomics: digging deeper with complete mitochondrial genomes. Trends Ecol Evol 14:394–398
Rosenberg MS, Kumar S (2001) Incomplete taxon sampling is not a problem for phylogenetic inference. Proc Natl Acad Sci USA 98:10751–10756
Phillips MJ, Penny D (2003) The root of the mammalian tree inferred from whole mitochondrial genomes. Mol Phylogenet Evol 28:171–185
Harrison GL, McLenachan PA, Phillips MJ, Slack KE, Cooper A, Penny D (2004) Four new avian mitochondrial genomes help get to basic evolutionary questions in the late cretaceous. Mol Biol Evol 21:974–983
Pereira SL, Baker AJ (2006) A mitogenomic timescale for birds detects variable phylogenetic rates of molecular evolution and refutes the standard molecular clock. Mol Biol Evol 23:1731–1740
Slack KE, Delsuc F, McLenachan PA, Arnason U, Penny D (2007) Resolving the root of the avian mitogenomic tree by breaking up long branches. Mol Phylogenet Evol 42:1–13
Morgan-Richards M, Trewick SA, Bartosch-Harlid A, Kardailsky O, Phillips MJ, McLenachan PA, Penny D (2008) Bird evolution: testing the Metaves clade with six new mitochondrial genomes. BMC Evol Biol 8:20
Mindell DP, Sorenson MD, Dimcheff DE, Hasegawa M, Ast JC, Yuri T (1999) Interordinal relationships of birds and other reptiles based on whole mitochondrial genomes. Syst Biol 48:138–152
van Tuinen M, Sibley CG, Hedges SB (2000) The early history of modern birds inferred from DNA sequences of nuclear and mitochondrial ribosomal genes. Mol Biol Evol 17:451–457
Livezey BC, Zusi RL (2007) Higher-order phylogeny of modern birds (Theropoda, Aves: Neornithes) based on comparative anatomy: II. Analysis and discussion. Zool J Linn Soc 149:1–94
Donne-Gousse C, Laudet V, Hanni C (2002) A molecular phylogeny of Anseriformes based on mitochondrial DNA analysis. Mol Phylogenet Evol 23:339–356
Worthy TH, Lee MSY (2008) Affinities of Miocene waterfowl (Anatidae: Manuherikia, Dunstanetta and Miotadorna) from the St Bathans Fauna, New Zealand. Palaeontology 51:677–708
van Tuinen M, Hedges SB (2001) Calibration of avian molecular clocks. Mol Biol Evol 18:206–213
Weir JT, Schluter D (2008) Calibrating the avian molecular clock. Mol Ecol 17:2321–2328
Welch JJ, Bromham L (2005) Molecular dating when rates vary. Trends Ecol Evol 20:320–327
Thorne JL, Kishino H (2002) Divergence time and evolutionary rate estimation with multilocus data. Syst Biol 51:689–702
Azuma Y, Kumazawa Y, Miya M, Mabuchi K, Nishida M (2008) Mitogenomic evaluation of the historical biogeography of cichlids toward reliable dating of teleostean divergences. BMC Evol Biol 8:215
van Tuinen M, Dyke GJ (2004) Calibration of galliform molecular clocks using multiple fossils and genetic partitions. Mol Phylogenet Evol 30:74–86
Pereira SL, Baker AJ (2006) A molecular timescale for galliform birds accounting for uncertainty in time estimates and heterogeneity of rates of DNA substitutions across lineages and sites. Mol Phylogenet Evol 38:499–509
Sorenson MD, Quinn TW (1998) Numts: a challenge for avian systematics and population biology. Auk 115:214–221
Ewing B, Hillier L, Wendl MC, Green P (1998) Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res 8:175–185
Ewing B, Green P (1998) Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res 8:186–194
Gordon D, Abajian C, Green P (1998) Consed: a graphical tool for sequence finishing. Genome Res 8:195–202
Wyman SK, Jansen RK, Boore JL (2004) Automatic annotation of organellar genomes with DOGMA. Bioinformatics 20:3252–3255
Slack KE, Janke A, Penny D, Arnason U (2003) Two new avian mitochondrial genomes (penguin and goose) and a summary of bird and reptile mitogenomic features. Gene 302:43–52
Feinstein J (2006) The mitochondrial genome of Cygnus columbianus, the Whistling Swan. DNA Seq 17:99–106
Lowe TM, Eddy SR (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25:955–964
Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R (2003) DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19:2496–2497
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Posada D, Crandall KA (1998) MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817–818
Nylander JA, Ronquist F, Huelsenbeck JP, Nieves-Aldrey JL (2004) Bayesian phylogenetic analysis of combined data. Syst Biol 53:47–67
Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Yang Z (1997) PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci 13:555–556
Haddrath O, Baker AJ (2001) Complete mitochondrial DNA genome sequences of extinct birds: ratite phylogenetics and the vicariance biogeography hypothesis. Proc Biol Sci 268:939–945
Clarke JA, Tambussi CP, Noriega JI, Erickson GM, Ketcham RA (2005) Definitive fossil evidence for the extant avian radiation in the Cretaceous. Nature 433:305–308
Rodriguez-Trelles F, Tarrio R, Ayala FJ (2002) A methodological bias toward overestimation of molecular evolutionary time scales. Proc Natl Acad Sci USA 99:8112–8115
Desjardins P, Morais R (1990) Sequence and gene organization of the chicken mitochondrial genome. A novel gene order in higher vertebrates. J Mol Biol 212:599–634
Nishibori M, Hayashi T, Tsudzuki M, Yamamoto Y, Yasue H (2001) Complete sequence of the Japanese quail (Coturnix japonica) mitochondrial genome and its genetic relationship with related species. Anim Genet 32:380–385
Nishibori M, Tsudzuki M, Hayashi T, Yamamoto Y, Yasue H (2002) Complete nucleotide sequence of the Coturnix chinensis (blue-breasted quail) mitochondrial genome and a phylogenetic analysis with related species. J Hered 93:439–444
Nishibori M, Hayashi T, Yasue H (2004) Complete nucleotide sequence of Numida meleagris (Helmeted Guineafowl) mitochondrial genome. J Poult Sci 41:259–268
Nishibori M, Shimogiri T, Hayashi T, Yasue H (2005) Molecular evidence for hybridization of species in the genus Gallus except for Gallus varius. Anim Genet 36:367–375
Guan X, Silva P, Gyenai KB, Xu J, Geng T, Tu Z, Samuels DC, Smith EJ (2009) The mitochondrial genome sequence and molecular phylogeny of the turkey, Meleagris gallopavo. Anim Genet 40:134–141
Ojala D, Merkel C, Gelfand R, Attardi G (1980) The tRNA genes punctuate the reading of genetic information in human mitochondrial DNA. Cell 22:393–403
Ojala D, Montoya J, Attardi G (1981) tRNA punctuation model of RNA processing in human mitochondria. Nature 290:470–474
Saccone C, De Giorgi C, Gissi C, Pesole G, Reyes A (1999) Evolutionary genomics in Metazoa: the mitochondrial DNA as a model system. Gene 238:195–209
Meiklejohn CD, Montooth KL, Rand DM (2007) Positive and negative selection on the mitochondrial genome. Trends Genet 23:259–263
Saccone C, Pesole G, Sbisa E (1991) The main regulatory region of mammalian mitochondrial DNA: structure–function model and evolutionary pattern. J Mol Evol 33:83–91
Ruokonen M, Kvist L (2002) Structure and evolution of the avian mitochondrial control region. Mol Phylogenet Evol 23:422–432
Ghivizzani SC, Madsen CS, Nelen MR, Ammini CV, Hauswirth WW (1994) In organello footprint analysis of human mitochondrial DNA: human mitochondrial transcription factor A interactions at the origin of replication. Mol Cell Biol 14:7717–7730
Mindell DP, Sorenson MD, Dimcheff DE (1998) An extra nucleotide is not translated in mitochondrial ND3 of some birds and turtles. Mol Biol Evol 15:1568–1571
Beckenbach AT, Robson SK, Crozier RH (2005) Single nucleotide +1 frameshifts in an apparently functional mitochondrial cytochrome b gene in ants of the genus Polyrhachis. J Mol Evol 60:141–152
Jermin LS, Guaur D, Crozier RH (1995) Evidence from analyses of intergenic regions for strand-specific directional mutation pressure in metazoan mitochondrial DNA. Mol Biol Evol 15:558–563
Reyes A, Gissi C, Pesole G, Saccone C (1998) Asymmetrical directional mutation pressure in the mitochondrial genome of mammals. Mol Biol Evol 15:957–966
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376
Brown JW, Rest JS, Garcia-Moreno J, Sorenson MD, Mindell DP (2008) Strong mitochondrial DNA support for a Cretaceous origin of modern avian lineages. BMC Biol 6:6
He L, Dai B, Zeng B, Zhang X, Chen B, Yue B, Li J (2009) The complete mitochondrial genome of the Sichuan Hill Partridge (Arborophila rufipectus) and a phylogenetic analysis with related species. Gene 435:23–28
Ericson PG, Christidis L, Irestedt M, Norman JA (2002) Systematic affinities of the lyrebirds (Passeriformes: Menura), with a novel classification of the major groups of passerine birds. Mol Phylogenet Evol 25:53–62
Dyke GJ, Gulas BE, Crowe TM (2003) Suprageneric relationships of galliform birds (Aves, Galliformes): a cladistic analysis of morphological characters. Zool J Linn Soc 137:227–244
Kriegs JO, Matzke A, Churakov G, Kuritzin A, Mayr G, Brosius J, Schmitz J (2007) Waves of genomic hitchhikers shed light on the evolution of gamebirds (Aves: Galliformes). BMC Evol Biol 7:190
Sibley CG, Ahlquist JE (1990) Phylogeny and classification of birds: a study in molecular evolution. Yale University Press, New Haven
Livezey BC (1986) A phylogenetic analysis of recent anseriform genera using morphological characters. Auk 105:681–698
Del Hoyo J, Elliot A, Sargatal J (1992) Handbook of the birds of the world, vol 2. New world vultures to guineafowl. Lynx Edicions, Barcelona
Livezey BC (1997) A phylogenetic classification of waterfowl (Aves: Anseriformes), including selected fossil species. Ann Carnegie Mus 66:457–496
Mayr G, Weidig I (2004) The Early Eocene bird Gallinuloides wyomingensis—a stem group representative of Galliformes. Acta Palaeontol Pol 49:211–217
Mayr G (2006) New specimens of the early Eocene stem group galliform Paraortygoides (Gallinuloididae), with comments on the evolution of a crop in the stem lineage of Galliformes. J Ornithol 147:31–37
Cracraft J (2001) Avian evolution, Gondwana biogeography and the Cretaceous-Tertiary mass extinction event. Proc Biol Sci 268:459–469
Pereira SL, Johnson KP, Clayton DH, Baker AJ (2007) Mitochondrial and nuclear DNA sequences support a Cretaceous origin of Columbiformes and a dispersal-driven radiation in the Paleocene. Syst Biol 56:656–672
Tavares ES, Baker AJ, Pereira SL, Miyaki CY (2006) Phylogenetic relationships and historical biogeography of neotropical parrots (Psittaciformes: Psittacidae: Arini) inferred from mitochondrial and nuclear DNA sequences. Syst Biol 55:454–470
Groth JG, Barrowclough GF (1999) Basal divergences in birds and the phylogenetic utility of the nuclear RAG-1 gene. Mol Phylogenet Evol 12:115–123
Acknowledgements
We gratefully express our thanks to Sergio L. Pereira for his kind help on molecular dating analyses. We also thank Mengjie Qiu and Jennifer Li for DNA sequencing, Jianfeng Ren and Xin Shen for useful discussion and Matt Phillips for help with phylogenetic analyses.
Author information
Authors and Affiliations
Corresponding author
Additional information
Feng Jiang and Yongwang Miao contributed equally to this work.
Rights and permissions
About this article
Cite this article
Jiang, F., Miao, Y., Liang, W. et al. The complete mitochondrial genomes of the whistling duck (Dendrocygna javanica) and black swan (Cygnus atratus): dating evolutionary divergence in Galloanserae. Mol Biol Rep 37, 3001–3015 (2010). https://doi.org/10.1007/s11033-009-9868-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-009-9868-9