Abstract
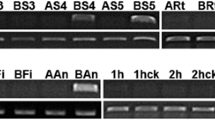
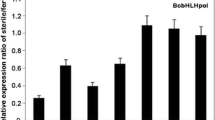
Two transcript-derived fragments (GenBank accession number DN237907.1 and DN237908.1) with high homology accumulated in the wild-type flower buds of Chinese cabbage (Brassica rapa L. ssp. chinensis Makino) are isolated and investigated. By rapid amplification of cDNA ends (RACE), the full length cDNA of the two fragments were obtained. The alignment of their cDNA sequence showed that they are identical except for differences in a few nucleotides and should belong to the same gene, namely, B rassica rapa M ale F ertile 5 (BcMF5). The BcMF5 gene consists of 252 bp encoding a protein of 83 amino acids and is interrupted by an intron of 256 bp. Sequence blast analysis revealed that BcMF5 is a member of the pollen coat protein (PCP) gene family and shared a high homology to SLR-BP. In the process of 3′RACE, eight different lengths of 3′-UTR sequence are found from the wild type of the mmc mutant. Southern blot analysis showed that BcMF5 could be a single-copy gene in the Chinese cabbage genome, implying that eight different lengths of 3′-UTR sequences might come from the same gene and could be a result of multiple sites polyadenylation of 3′-UTRs of BcMF5. Based on sequence analysis, southern hybridization combined with RT-PCR, and northern hybridization, it was discovered that 3′-UTRs of BcMF5 contained some functional elements and their temporal and spatial expression patterns were different, but all strongly expressed in the stage IV and stage V flower buds of wild type. This indicate that different lengths of 3′-UTR may be involved in a regulation mechanism during the transcription of BcMF5.







Similar content being viewed by others
Abbreviations
- BcMF5:
-
Brassica rapa Male Fertile 5
- PCP:
-
Pollen coat protein
References
Ye WZ, Cao JS, Xiang X, Zeng GW (2003) Molecular cloning and characterization of the genic male sterility related gene CYP86MF in Chinese cabbage (Brassica campestris L. ssp. chinensis Makino var. communis Tsen et Lee). J Hort Sci Biotechnol 78:319–323
Yu XL, Cao JS, Ye WZ, Wang YQ (2004) Construction of an antisense CYP86MF gene plasmid vector and production of a male-sterile Chinese cabbage transformant by the pollen-tube method. J Hort Sci Biotechnol 79:833–839
Cao JS, Yu XL, Ye WZ, Lu G, Xiang X (2006) Functional analysis of a novel male fertility CYP86MF gene in Chinese cabbage (Brassica campestris L. ssp. chinensismakino). Plant Cell Rep 24:715–723
Wang YQ, Ye WZ, Cao JS, Yu XL, Xiang X, Lu G (2005) Cloning and characterization of the microspore development related gene BcMF2 in Chinese cabbage pak-choi (Brassica campestris L. ssp. chinensis Makino). J Integr Plant Biol 8:4763–872
Wang YQ, Yu XL, Cao JS (2004) Isolation and characterization of BcMF3, a gene expressed only in maintainer line in Chinese cabbage-pak-choi (Brassica campestris L. ssp. chinensis Makino var. communis Tsen et lee. Acta Genet Sin 31(11):1302–1308
Liu LC, Xiang X, Cao JS (2006) BcMF4 gene, encoding a lucine-rich repeat protein, plays a role in male fertility in Chinese cabbage-pak-choi. Hereditas 28(11):1428–1434
Chen DH, Ronald PC (1999) A rapid DNA minipreparation method suitable for AFLP and other PCR applications. Plant Mol Biol Rep 17:53–57
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Hiscock SJ, Doughty J, Willis AC, Dickinson HG (1995) A 7-kDa pollen coating-borne peptide from Brassica napus interacts with S-locus glycoprotein and S-locus related glycoprotein. Planta 196:367–374
Takayama S, Shiba H, Iwano M, Asano K, Hara M, Che F-S, Watanabe M, Hinata K, Isogai A (2000a) Isolation and characterization of pollen coat proteins of Brassica rapa that interact with S locus-related glycoprotein 1 involved in pollen-stigma adhesion. Proc Natl Acad Sci USA 97(7):3765–3770
Pesole G, Liuni S (1999) Internet resources for the functional analysis of 5’ and 3’ untranslated regions of eukaryotic mRNA. Trends Genet 15:379–380
Doughty J, Hedderson F, McCubbin A, Dickinson HG (1993) Interaction between a coating-borne peptide of the Brassica pollen grain and S (incompatibility)-locus linked stigmatic glycoproteins. Proc Natl Acad Sci USA90:467–471
Doughty J, Dixon S, Hiscock SH, Willis AC, Parkin IA Dickinson HG (1998) PCP-A1, a defensin-like Brassica pollen coat protein that binds the S locus glycoprotein, is the product of gametophytic gene expression. Plant Cell 10:1333–1348
Doughty J, Wong HY, Dickinson HG (2000) Cysteine-rich pollen coat proteins (PCPs) and their interactions with stigmatic S (incompatibility) and S-related proteins in Brassica: putative roles in SI and pollination. Ann Bot 85:161–169
Willing RP, Bashe D, Mascarenhas JP (1988) An analysis of the quantity and diversity of messenger RNAs from pollen and shoots of Zea mays. Theor Appl Genet 75: 751–753
Willing RP, Mascarenhas JP (1984) Analysis of the complexity and diversity of mRNAs from pollen and shoots of Tradescantia. Plant Physiol 75:865–868
Lord EM (2003) Adhesion and guidance in compatible pollination. J Exp Bot 54:47–54
Jiang YQ, Ling Y, Zhao WL (2001) Progress in the Studies on Function of 3′ Untranslated Region on Post-transcriptional Level. Chin Bull Bot 18(1):3–10
Jin YF, Bian TF (2004) Isolation and partial characterization of a novel pollen-specific cDNA with multiple polyadenylation sites from wheat. Acta Biochim Biophys Sin 36(7):467–476
Chen CYA, Shyu AB (1995) AU_richelements: characterization and importance in mRNA degradation. TIBS 20(11):465–470
Di Noia JM, D’ OrsoI, Sanchez DO, Frasch AC (2000) AU_rich elements in the 3’ untranslated region of a new mucin_type Gene family of Trypanosomacruzi confers mRNA instability and modulates translation efficiency. J Biol Chem 275:10218–10227
Acknowledgments
This work was supported by the Natural Science Foundation of China (No. 30671426) and the Key Sci-technology Project of Zhejiang Province (No. 2005C12019-02).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhang, Q., Cao, J., Huang, L. et al. BcMF5, a pollen coat protein gene (PCP), from Brassica rapa. ssp. chinensis, involved in the transcription of different lengths of 3′-UTRs of PCPs. Mol Biol Rep 35, 439–445 (2008). https://doi.org/10.1007/s11033-007-9104-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-007-9104-4