Abstract
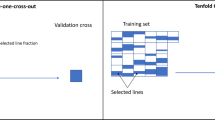
Agronomically important traits generally have complex genetic architecture, where many genes have a small and largely additive effect. Genomic prediction has been demonstrated to increase genetic gain and efficiency in plant breeding programs beyond marker-assisted selection and phenotypic selection. The objective of this study was to evaluate the impact of allelic origin, marker density, training population size, and cross-validation schemes on the accuracy of genomic prediction models in an interspecific soybean nested association mapping (NAM) panel. Three cross-validation schemes were used: (a) Within-Family (WF): training population and predictions are made exclusively within each family; (b) Across All families (AF): all the individuals from the three families were randomly assigned to either the training or validation set; (c) Leave one Family out (LFO): each family is predicted using a training set that contains the other two families. Predictive abilities increased with training population size up to 350 individuals, but no significant gains were noted beyond 250 individuals in the training population. The number of markers had a limited impact on the observed predictive ability across traits; increasing markers used in the model above 1000 revealed no significant increases in prediction accuracy. Predictive abilities for AF were not significantly different from the WF method, and predictive abilities across populations for the WF method had a range of 0.58 to 0.70 for maturity, protein, meal, and oil. Our results also showed encouraging prediction accuracies for grain yield (0.58–0.69) using the WF method. Partitioning genomic prediction between G. max and G. soja alleles revealed useful information to select material with a larger allele contribution from both parents and could accelerate allele introgression from exotic germplasm into the elite soybean gene pool.






Similar content being viewed by others
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Abdel-Haleem H, Ji P, Boerma HR, Li Z (2013) An R package for SNP marker-based parent-offspring tests. Plant Methods 9:44. https://doi.org/10.1186/1746-4811-9-44
Akdemir D, Sanchez JI, Jannink JL (2015) Optimization of genomic selection training populations with a genetic algorithm. Genet Sel Evol 47:38. https://doi.org/10.1186/s12711-015-0116-6
Allier A, Moreau L, Charcosset A, Teyssèdre S, Lehermeier C (2019) Usefulness criterion and post-selection parental contributions in multi-parental crosses: application to polygenic trait introgression. G3 Genes Genomes, Genet 9:1469–1479. https://doi.org/10.1534/g3.119.400129
Allier A, Teyssèdre S, Lehermeier C, Charcosset A, Moreau L (2020) Genomic prediction with a maize collaborative panel: identification of genetic resources to enrich elite breeding programs. Theor Appl Genet 133:201–215. https://doi.org/10.1007/s00122-019-03451-9
Arruda MP, Lipka AE, Brown PJ, Krill AM, Thurber C, Brown-Guedira G, Dong Y, Foresman BJ, Kolb FL (2016) Comparing genomic selection and marker-assisted selection for Fusarium head blight resistance in wheat (Triticum aestivum L.). Mol Breed 36:1–11. https://doi.org/10.1007/s11032-016-0508-5
Bates D, Mächler M, Bolker B, Walker S (2015) Fitting linear mixed-effects models using lme4. J Stat Softw 67:1–48. https://doi.org/10.18637/jss.v067.i01
Beche E, Gillman JD, Song Q, Nelson R, Beissinger T, Decker J, Shannon G, Scaboo AM (2020) Nested association mapping of important agronomic traits in three interspecific soybean populations. Theor Appl Genet 133:. doi: https://doi.org/10.1007/s00122-019-03529-4
Bernardo R, Yu J (2007) Prospects for Genomewide Selection for Quantitative Traits in Maize. Crop Science 47 (3):1082–1090. https://doi.org/10.2135/cropsci2006.11.0690
Bernard RL, Cremeens CR (1988) Registration of ‘Williams 82’ soybean. Crop Sci 28:1027–1028. https://doi.org/10.2135/cropsci1988.0011183X002800060049x
Berro I, Lado B, Nalin RS, Quincke M, Gutiérrez L (2019) Training population optimization for genomic selection. Plant Genome 12:190028. https://doi.org/10.3835/plantgenome2019.04.0028
Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23:2633–2635.https://doi.org/10.1093/bioinformatics/btm308
Calus MPL (2010) Genomic breeding value prediction: methods and procedures. Animal 4:157–164. https://doi.org/10.1017/S1751731109991352
Chung J, Babka HL, Graef GL, Staswick PE, Lee DJ, Cregan PB, Shoemaker RC, Specht JE (2003) The seed protein, oil, and yield QTL on soybean linkage group I. Crop Sci 43:1053–1067. https://doi.org/10.2135/cropsci2003.1053
Combs E, Bernardo R (2013) Genomewide selection to introgress semidwarf maize germplasm into U.S. Corn Belt inbreds. Crop Sci 53:1427–1436. https://doi.org/10.2135/cropsci2012.11.0666
Crossa J, Jarquín D, Franco J, et al (2016) Genomic prediction of Gene Bank Wheat Landraces. G3:Genes|Genomes|Genetics 6:1819–1834. doi: https://doi.org/10.1534/g3.116.029637
De Azevedo PL, Moellers TC, Zhang J et al (2017) Leveraging genomic prediction to scan germplasm collection for crop improvement. PLoS One 12:e0179191. https://doi.org/10.1371/journal.pone.0179191
De Los Campos G, Naya H, Gianola D et al (2009) Predicting quantitative traits with regression models for dense molecular markers and pedigree. Genetics 182:375–385. https://doi.org/10.1534/genetics.109.101501
de los Campos G, Sorensen D, Gianola D (2015) Genomic heritability: what is it? PLoS Genet 11:1–21. https://doi.org/10.1371/journal.pgen.1005048
Doebley JF, Gaut BS, Smith BD (2006) The molecular genetics of crop domestication. Cell 127:1309–1321. https://doi.org/10.1016/j.cell.2006.12.006
Duhnen A, Gras A, Teyssèdre S, Romestant M, Claustres B, Daydé J, Mangin B (2017) Genomic selection for yield and seed protein content in soybean: a study of breeding program data and assessment of prediction accuracy. Crop Sci 57:1325–1337. https://doi.org/10.2135/cropsci2016.06.0496
Edwards SM, Buntjer JB, Jackson R, et al (2019) The effects of training population design on genomic prediction accuracy in wheat. Theor Appl Genet 132:1943–1952. https://doi.org/10.1007/s00122-019-03327-y
Endelman JB (2011) Ridge regression and other kernels for genomic selection with R package rrBLUP. Plant Genome J 4:250–255. https://doi.org/10.3835/plantgenome2011.08.0024
FAOSTAT (2020) Food and Agriculture Organization of the United Nations Statistical Database. http://www.fao.org/faostat/en/#data/QC. Accessed 13 Mar 2020
Fehr WR (1991) Principles of cultivar development. Vol. 1. Theory and technique. Macmillan, New York
Fehr WR, Caviness CE, Burmood DT, Pennington JS (1971) Stage of development descriptions for soybeans, Glycine max (L.) Merrill1. Crop Sci 11:929–931. https://doi.org/10.2135/cropsci1971.0011183X001100060051x
Gesteira G d S, Bruzi AT, Zito RK et al (2018) Selection of early soybean inbred lines using multiple indices. Crop Sci 58:2494–2502. https://doi.org/10.2135/cropsci2018.05.0295
Gizlice Z, Carter TE, Burton JW (1994) Genetic base for North American public soybean cultivars released between 1947 and 1988. Crop Sci 34:1143–1151. https://doi.org/10.2135/cropsci1994.0011183X003400050001x
Gorjanc G, Jenko J, Hearne SJ, Hickey JM (2016) Initiating maize pre-breeding programs using genomic selection to harness polygenic variation from landrace populations. BMC Genomics 17:30. https://doi.org/10.1186/s12864-015-2345-z
Habier D, Fernando RL, Dekkers JCM (2007) The impact of genetic relationship information on genome-assisted breeding values. Genetics 177:2389–2397. https://doi.org/10.1534/genetics.107.081190
Heffner EL, Lorenz AJ, Jannink JL, Sorrells ME (2010) Plant breeding with genomic selection: gain per unit time and cost. Crop Sci 50:1681–1690. https://doi.org/10.2135/cropsci2009.11.0662
Heffner EL, Jannink J-L, Sorrells ME (2011) Genomic selection accuracy using multifamily prediction models in a wheat breeding program. Plant Genome 4:65–75. https://doi.org/10.3835/plantgenome2010.12.0029
Hegstad JM, Nelson RL, Renny-Byfield S, Feng L, Chaky JM (2019) Introgression of novel genetic diversity to improve soybean yield. Theor Appl Genet 132:2541–2552. https://doi.org/10.1007/s00122-019-03369-2
Hill WG (2010) Understanding and using quantitative genetic variation. Philos Trans R Soc B Biol Sci 365:73–85. https://doi.org/10.1098/rstb.2009.0203
Huang BE, George AW, Forrest KL, Kilian A, Hayden MJ, Morell MK, Cavanagh CR (2012) A multiparent advanced generation inter-cross population for genetic analysis in wheat. Plant Biotechnol J 10:826–839. https://doi.org/10.1111/j.1467-7652.2012.00702.x
Hyten DL, Song Q, Zhu Y, Choi IY, Nelson RL, Costa JM, Specht JE, Shoemaker RC, Cregan PB (2006) Impacts of genetic bottlenecks on soybean genome diversity. Proc Natl Acad Sci U S A 103:16666–16671. https://doi.org/10.1073/pnas.0604379103
Isidro J, Jannink JL, Akdemir D, Poland J, Heslot N, Sorrells ME (2015) Training set optimization under population structure in genomic selection. Theor Appl Genet 128:145–158. https://doi.org/10.1007/s00122-014-2418-4
Jannink JL (2010) Dynamics of long-term genomic selection. Genet Sel Evol 42:35. https://doi.org/10.1186/1297-9686-42-35
Jarquín D, Kocak K, Posadas L, Hyma K, Jedlicka J, Graef G, Lorenz A (2014) Genotyping by sequencing for genomic prediction in a soybean breeding population. BMC Genomics 15:740. https://doi.org/10.1186/1471-2164-15-740
Jarquin D, Specht J, Lorenz A (2016) Prospects of genomic prediction in the USDA soybean germplasm collection: historical data creates robust models for enhancing selection of accessions. G3 genes. Genomes, Genet 6:2329–2341. https://doi.org/10.1534/g3.116.031443
La T, Large E, Taliercio E et al (2019) Characterization of select wild soybean accessions in the USDA germplasm collection for seed composition and agronomic traits. Crop Sci 59:1–19. https://doi.org/10.2135/cropsci2017.08.0514
Liu B, Fujita T, Yan Z-H, Sakamoto S, Xu D, Abe J (2007) QTL mapping of domestication-related traits in soybean (Glycine max). Ann Bot 100:1027–1038. https://doi.org/10.1093/aob/mcm149
Lorenz AJ (2013) Resource allocation for maximizing prediction accuracy and genetic gain of genomic selection in plant breeding: a simulation experiment. G3:Genes|Genomes|Genetics 3:481–491. https://doi.org/10.1534/g3.112.004911
Lorenz AJ, Smith KP (2015) Adding genetically distant individuals to training populations reduces genomic prediction accuracy in barley. Crop Sci 55:2657–2667. https://doi.org/10.2135/cropsci2014.12.0827
Meuwissen THE, Hayes BJ, Goddard ME (2001) Prediction of total genetic value using genome-wide dense marker maps. Genetics 157:1819–1829
Michel S, Löschenberger F, Ametz C, Pachler B, Sparry E, Bürstmayr H (2019) Simultaneous selection for grain yield and protein content in genomics-assisted wheat breeding. Theoretical and Applied Genetics 132(6):1745–1760. Selection for Quantitative. https://doi.org/10.1007/s00122-019-03312-5
Norman A, Taylor J, Edwards J, Kuchel H (2018) Optimising genomic selection in wheat: effect of marker density, population size and population structure on prediction accuracy. G3:Genes|Genomes|Genetics 8:2889–2899. doi: https://doi.org/10.1534/g3.118.200311
Pantalone V, Smallwood C (2018) Registration of ‘TN11-5102’ soybean cultivar with high yield and high protein meal. J Plant Regist 12:304–308. https://doi.org/10.3198/jpr2017.10.0074crc
Remington DL, Thornsberry JM, Matsuoka Y, Wilson LM, Whitt SR, Doebley J, Kresovich S, Goodman MM, Buckler ES (2001) Structure of linkage disequilibrium and phenotypic associations in the maize genome. Proc Natl Acad Sci U S A 98:11479–11484. https://doi.org/10.1073/pnas.201394398
Rincent R, Laloë D, Nicolas S, Altmann T, Brunel D, Revilla P, Rodríguez VM, Moreno-Gonzalez J, Melchinger A, Bauer E, Schoen CC, Meyer N, Giauffret C, Bauland C, Jamin P, Laborde J, Monod H, Flament P, Charcosset A, Moreau L (2012) Maximizing the reliability of genomic selection by optimizing the calibration set of reference individuals: comparison of methods in two diverse groups of maize inbreds (Zea mays L.). Genetics 192:715–728. https://doi.org/10.1534/genetics.112.141473
Rincent R, Charcosset A, Moreau L (2017) Predicting genomic selection efficiency to optimize calibration set and to assess prediction accuracy in highly structured populations. Theor Appl Genet 130:2231–2247. https://doi.org/10.1007/s00122-017-2956-7
Rincker K, Nelson R, Specht J, Sleper D, Cary T, Cianzio SR, Casteel S, Conley S, Chen P, Davis V, Fox C, Graef G, Godsey C, Holshouser D, Jiang GL, Kantartzi SK, Kenworthy W, Lee C, Mian R, McHale L, Naeve S, Orf J, Poysa V, Schapaugh W, Shannon G, Uniatowski R, Wang D, Diers B (2014) Genetic improvement of U.S. soybean in maturity groups II, III, and IV. Crop Sci 54:1419–1432. https://doi.org/10.2135/cropsci2013.10.0665
Ru S, Bernardo R (2020) Predicted genetic gains from introgressing chromosome segments from exotic germplasm into an elite soybean cultivar. Theor Appl Genet 133:605–614. https://doi.org/10.1007/s00122-019-03490-2
Song Q, Yan L, Quigley C, et al (2020) Soybean BARCSoySNP6K-an assay for soybean genetics and breeding research. Plant J TPJ.14960. doi: https://doi.org/10.1111/tpj.14960
Stewart-Brown BB, Song Q, Vaughn JN, Li Z (2019) Genomic selection for yield and seed composition traits within an applied soybean breeding program. G3 Genes, Genomes, Genet 9:2253–2265. https://doi.org/10.1534/g3.118.200917
University of Missouri Extension (1993) Soybean production in Missouri. https://extension.missouri.edu/g4410#date. Accessed 27 Oct 2020
Xavier A, Muir WM, Rainey KM (2016) Assessing predictive properties of genome-wide selection in soybeans. G3:Genes|Genomes|Genetics 6:2611–6. doi: https://doi.org/10.1534/g3.116.032268
Xavier A, Muir WM, Rainey KM (2019) bWGR: Bayesian whole-genome regression. Bioinformatics 36:1957–1959. https://doi.org/10.1093/bioinformatics/btz794
Yang CJ, Sharma R, Gorjanc G, et al (2020) Origin specific genomic selection: a simple process to optimize the favorable contribution of parents to progeny. G3 (Bethesda) 10:2445–2455. doi: https://doi.org/10.1534/g3.120.401132
Acknowledgements
The authors would like to acknowledge the personnel from the soybean breeding program at the University of Missouri for their time and effort in preparing and conducting the field experiments.
Code availability
Not applicable
Funding
This research was funded by the Missouri Soybean Merchandising Council and the United Soybean Board.
Author information
Authors and Affiliations
Contributions
EB conducted field evaluations and data analysis; AS acquired funding and supervised the research; QS performed the genotyping; RN developed the initial populations; EB and AS wrote the paper; JG, TB, JD, GS, RN, and QS revised and edited the manuscript. All authors read the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
ESM 1
(DOCX 1493 kb)
Rights and permissions
About this article
Cite this article
Beche, E., Gillman, J.D., Song, Q. et al. Genomic prediction using training population design in interspecific soybean populations. Mol Breeding 41, 15 (2021). https://doi.org/10.1007/s11032-021-01203-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-021-01203-6