Abstract
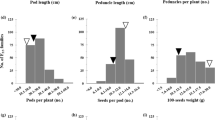
Yardlong bean [Vigna unguiculata (L.) Walp. ssp. unguiculata cv.-gr. sesquipedalis] is a vegetable legume crop evolved from cultivated grain cowpea (V. unguiculata ssp. unguiculata cv.-gr. unguiculata) which is domesticated from wild cowpea. It has a dramatic change in pod length and pod fiber content, and a complete loss of pod shattering, as compared to its wild progenitor. In this study, we identified quantitative trait loci (QTLs) controlling pod fiber content and pod shattering in two populations (BC1F1 and F2) derived from a cross between yardlong bean and wild cowpea. BC1F1:2 and F2:3 families were grown under field condition in which insoluble dietary fiber (cellulose, hemicelluloses and lignin) contents in mature pods, and pod shattering were evaluated. Correlation analysis showed positive relationship among the types of fiber, and between the fiber and shattering. Inclusive composite interval mapping revealed that a major QTL on linkage group 7 (LG7) controlled cellulose, hemicellulose and lignin contents in pod, and pod shattering. The other QTLs related to pod fibers on LG1 and LG4 also co-localized with the QTLs for pod shattering. Comparative genome analysis with azuki bean (Vigna angularis) suggested that the QTL region on LG7 for cellulose, hemiclellulose, lignin and pod shattering in yardlong bean contains genes encoding MYB transcription factor, MYB83, regulating biosynthesis of the three fibers, while the QTL region for cellulose and shattering of pod on LG1 harbors gene encoding cellulose synthase A7 (CESA7). These genes may be important targets for functional study to reveal major factors regulating pod fiber biosynthesis and pod shattering in yardlong bean.



Similar content being viewed by others
References
Andargie M, Pasquet RS, Gowda BS, Muluvi GM, Timko MP (2014) Molecular mapping of QTLs for domestication-related traits in cowpea (V. unguiculata (L.) Walp.). Euphytica 200:401–412
Bonawitz ND, Chapple C (2010) The genetics of lignin biosynthesis: connecting genotype to phenotype. Annu Rev Genet 44:337–363
Dong Y, Yang X, Liu J, Wang BH, Liu BL, Wang YZ (2014) Pod shattering resistance associated with domestication is mediated by a NAC gene in soybean. Nat Commun 5:3352. doi:10.1038/ncomms4352
Funatsuki H, Suzuki M, Hirose A, Inaba H, Yamada T, Hajika M, Komatsu K, Katayama T, SayamaT Ishimoto M, Fujino K (2014) Molecular basis of a shattering resistance boosting global dissemination of soybean. Proc Natl Acad Sci USA 111:17797–17802
Gepts P, Debouck DG (1991) Origin, domestication, and evolution of the common bean, Phaseolus vulgaris. In: Voysest O, Van Schoonhoven A (eds) Common beans: research for crop improvement. CAB, Oxon, pp 7–53
Gioia T, Logozzo G, Kami J, Spagnoletti-Zeuli P, Gepts P (2013) Identification and characterization of a homologue to the Arabidopsis INDEHISCENT gene in common bean. J Hered 104:273–286
Hussey SG, Mizrachi E, Creux NM, Myburg AA (2013) Navigating the transcriptional roadmap regulating plant secondary cell wall deposition. Front Plant Sci 4:325. doi:10.3389/fpls.2013.00325
Isemura T, Kaga A, Konishi S, Ando T, Tomooka N, Han OK, Vaughan DA (2007) Genome dissection of traits related to domestication in azuki bean (Vigna angularis) and their comparison with other warm season legumes. Ann Bot 100:1053–1071
Isemura T, Kaga A, Tomooka N, Ishimizu T, Vaughan DA (2010) The genetics of domestication of rice bean, Vigna umbellata. Ann Bot 106:927–944
Isemura T, Kaga A, Tabata S, Somta P, Srinives P, Shimizu T et al (2012) Construction of a genetic linkage map and genetic analysis of domestication related traits in mungbean (Vigna radiata). PLoS ONE 7(8):e41304. doi:10.1371/journal.pone.0041304
Kang YJ, Kim SK, Kim MY, Lestari P, Kim KH, Ha BK et al (2014) Genome sequence of mungbean and insights into evolution within Vigna species. Nat Commun 5:5443. doi:10.1038/ncomms6443
Kang YJ, Satyawan D, Shim S, Lee T, Lee J, Hwang WJ et al (2015) Draft genome sequence of adzuki bean, Vigna angularis. Sci Rep 5:8069. doi:10.1038/srep08069
Ko JH, Kim WC, Han KH (2009) Ectopic expression of MYB46 identifies transcriptional regulatory genes involved in secondary wall biosynthesis in Arabidopsis. Plant J 60:649–665
Ko JH, Jeon HW, Kim WC, Kim JY, Han KH (2014) The MYB46/MYB83-mediated transcriptional regulatory programme is a gatekeeper of secondary wall biosynthesis. Ann Bot 114:1099–1107
Koinange EM, Singh SP, Gepts P (1996) Genetic control of the domestication syndrome in common bean. Crop Sci 36:1037–1045
Kongjaimun A, Kaga A, Tomooka N, Somta P, Shimizu T, Shu Y et al (2012a) An SSR-based linkage map of yardlong bean (Vigna unguiculata (L.) Walp. subsp. unguiculata sesquipedalis group) and QTL analysis of pod length. Genome 55:81–92
Kongjaimun A, Kaga A, Tomooka N, Somta P, Vaughan DA, Srinives P (2012b) The genetics of domestication of yardlong bean, Vigna unguiculata (L.) Walp. ssp. unguiculata cv.-gr. sesquipedalis. Ann Bot 109:1185–1200
Kongjaimun A, Somta P, Tomooka N, Kaga A, Vaughan DA, Srinives P (2013) QTL mapping of pod tenderness and total soluble solid in yardlong bean [Vigna unguiculata (L.) Walp. subsp. unguiculata cv.-gr. sesquipedalis]. Euphytica 189:217–223
Liljegren SJ, Ditta GS, Eshed Y, Savidge B, Bowman J, Yanofsky MF (2000) SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature 404:766–770
Liljegren SJ, Roeder AH, Kempin SA, Gremski K, Østergaard L, Guimil S et al (2004) Control of fruit patterning in Arabidopsis by INDEHISCENT. Cell 116:843–853
Lush WM, Evans LT (1981) The domestication and improvement of cowpeas (Vigna unguiculata (L.) Walp.). Euphytica 30:579–587
Maréchal R, Mascherpa JM, Stainier F (1978) Etude taxonomique d’un groupe complex d’espèces des geners Phaseolus et Vigna (Papilionaceae) sur la base de données morphologiques et polliniques, traitées, par l’analyse informatique. Boisseira 28:1–273
McCarthy RL, Zhong R, Ye ZH (2009) MYB83 is a direct target of SND1 and acts redundantly with MYB46 in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell Physiol 50:1950–1964
McFarlane HE, Döring A, Persson S (2014) The cell biology of cellulose synthesis. Annu Rev Plant Biol 65:69–94
Nanni L, Bitocchi E, Bellucci E, Rossi M, Rau D, Attene G, Gepts P, Papa R (2011) Nucleotide diversity of a genomic sequence similar to SHATTERPROOF (PvSHP1) in domesticated and wild common bean (Phaseolus vulgaris L.). Theor Appl Genet 123:1341–1357
R Development Core Team (2012) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna
Rippert P, Puyaubert J, Grisollet D, Derrier L, Matringe M (2009) Tyrosine and phenylalanine are synthesized within the plastids in Arabidopsis. Plant Physiol 149:1251–1260
Romkaew J, Nagaya Y, Goto M, Suzuki K, Umezaki T (2008) Pod dehiscence in relation to chemical components of pod shell in soybean. Plant Prod Sci 11:278–282
Sakai H, Naito K, Ogiso-Tanaka E, Takahashi Y, Iseki K, Muto C et al (2015a) The power of single molecule real-time sequencing technology in the de novo assembly of a eukaryotic genome. Sci Rep 5:16780. doi:10.1038/srep16780
Sakai H, Naito K, Takahashi Y, Sato T, Yamamoto T, Muto I, Itoh T, Tomooka N (2015b) The Vigna Genome Server, ‘VigGS’: a genomic knowledge base of the genus Vigna based on high quality, annotated genome sequence of the azuki bean, Vigna angularis (Willd.) Ohwi & Ohashi. Plant Cell. doi:10.1093/pcp/pcv189
Somta P, Musch W, Kongsamai B, Chanprame S, Nakasathien S, Toojinda T, Sorajjapinun W, Seehalak W, Tragoonrung S, Srinives P (2008) New microsatellite markers isolated from mungbean (Vigna radiata (L.) Wilczek). Mol Ecol Resour 8:1155–1157
Steele WM, Mehra KL (1980) Structure, evolution, and adaptation to farming systems and environments in Vigna. In: Summerfield RJ, Bunting AH (eds) Advances in legume sciences. Royal Botanic Gardens, Kew
Taylor NG, Scheible WR, Cutler S, Somerville CR, Turner SR (1999) The irregular xylem3 locus of Arabidopsis encodes a cellulose synthase required for secondary cell wall synthesis. Plant Cell 11:769–779
Temnykh S, DeClerck G, Lukashova A, Lipovich L, Cartinhour S, McCouch S (2001) Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): frequency, length variation, transposon associations, and genetic marker potential. Genome Res 11:1441–1452
Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG (2012) Primer3—new capabilities and interfaces. Nucl Acids Res 40:e115
Van Soest PJ, Robertson JB, Lewis BA (1991) Methodsfor dietary fiber, neutral detergent fiber, and nonstarchpolysaccharides in relation to animal nutrition. J Dairy Sci 74:3583–3597
Verdcourt B (1970) Studies in the Leguminosae-Papilionoideae for the ‘Flora of Tropical East Africa’: IV. Kew Bull 24:507–569
Vogel KP, Pedersen JF, Masterson SD, Toy JJ (1999) Evaluation of a filter bag system for NDF, ADF, and IVDMD forage analysis. Crop Sci 39:276–279
Wang S, Basten CJ, Zeng ZB (2012) Windows QTL cartographer 25. Department of Statistics, North Carolina State University, Raleigh
Zeng ZB (1994) Precision mapping of quantitative trait loci. Genetics 136:1457–1466
Zhong R, Ye Z-H (2015) Secondary cell walls: biosynthesis, patterned deposition and transcriptional regulation. Plant Cell Physiol 56:195–1214
Zhong R, Richardson EA, Ye Z-H (2007) The MYB46 transcription factor is a direct target of SND1 and regulates secondary wall biosynthesis in Arabidopsis. Plant Cell 19:2776–2792
Zhong R, Lee C, Ye Z-H (2010) Evolutionary conservation of the transcriptional network regulating secondary cell wall biosynthesis. Trends Plant Sci 15:625–631
Acknowledgments
This research was financially supported by 2016 Reseach Fund of Department of Agronomy, Faculty of Agriculture at Kamphaeng Saen, Kasetsart University and by Kasetsart University Research and Development Institute.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Suanum, W., Somta, P., Kongjaimun, A. et al. Co-localization of QTLs for pod fiber content and pod shattering in F2 and backcross populations between yardlong bean and wild cowpea. Mol Breeding 36, 80 (2016). https://doi.org/10.1007/s11032-016-0505-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-016-0505-8