Abstract
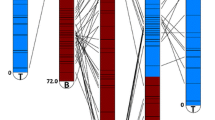
To facilitate marker-assisted breeding and analysis of the structure and/or organization of Capsicum (pepper) genomes, this study utilized expressed sequence tags (ESTs) to develop single-nucleotide polymorphism (SNP) markers. Three different types of PCR-based markers derived from pepper ESTs were developed: intron-based polymorphic markers (IBPs), conserved ortholog sets (COSIIs), and eSNPs (EST–SNPs). For scanning and detection of SNPs, high-resolution melting analysis was performed and the resultant markers were used for linkage analysis. A total of 512 markers, comprising 214 IBP, 143 COSII, 48 eSNP, and 107 previously reported markers, were mapped on 12 linkage groups (LGs) of the “AC99” F2 population. This newly constructed interspecific map (AC2) covered 2,335.6 cM with an average marker interval distance of 4.5 cM and was aligned directly with another interspecific map (AF) for validation. Most LGs showed collinear relationships, except for the alignment of chromosomes 1 and 8 of the AC2 map to LG P1 of the AF map. Using our newly developed SNP markers, we generated chromosome-specific markers, and the previously predicted reciprocal translocation event between chromosomes 1 and 8 was revealed between wild and cultivated Capsicum by fluorescent in situ hybridization analysis. The results from this study will promote subsequent evolutionary studies of Capsicum species.


Similar content being viewed by others
References
An SJ, Pandeya D, Park SW, Li J, Kwon JK, Koeda S, Hosokawa M, Paek NC, Choi D, Kang BC (2010) Characterization and genetic analysis of a low-temperature-sensitive mutant, sy-2, in Capsicum chinense. Theor Appl Genet 122:459–470
Ashirafi H, Hill T, Stoffel K, Kozik A, Yao J, Chin-Wo SR, Van Deynze A (2012) De novo assembly of the pepper transcriptome (Capsicum annuum): a benchmark for in silico discovery of SNPs, SSRs and candidate genes. BMC Genomics 13:571
Aza-González C, Núñez-Palenius HG, Ochoa-Alejo N (2011) Molecular biology of capsaicinoid biosynthesis in chilli pepper (Capsicum spp.). Plant Cell Rep 30:695–706
Ben-Chaim A, Borovsky Y, Falise M, Mazourek M, Kang BC, Paran I, Jahn M (2006) QTL analysis for capsaicinoid content in Capsicum. Theor Appl Genet 113:1481–1490
Chagné D, Gasic K, Crowhurst RN, Han Y, Bassett HC, Bowatte DR, Lawrence TJ, Rikkerink EH, Gardiner SE, Korban SS (2008) Development of a set of SNP markers present in expressed genes of the apple. Genomics 92:353–358
Hearnden PR, Eckermann PJ, McMichael GL, Hayden MJ, Eglinton JK, Chalmers KJ (2007) A genetic map of 1,000 SSR and DArT markers in a wide barley cross. Theor Appl Genet 115:383–391
Hong HT, Kim KT, Kim S, Chae Y, Park JH, Oh DG, Cho MC (2010) Comparative mapping of consensus SSR markers in an intraspecific F8 recombinant inbred line population in Capsicum. Hort Environ Biotechnol 51:193–206
Jeong HJ, Kwon JK, Pandeya D, Hwang J, Hoang NH, Bae JH, Kang BC (2011) A survey of natural and ethyl methane sulfonate-induced variations of eIF4E using high-resolution melting analysis in Capsicum. Mol Breeding 29:349–360
Jung JK, Park SW, Liu WY, Kang BC (2010) Discovery of single nucleotide polymorphism in Capsicum and SNP markers for cultivar identification. Euphytica 175:91–107
Kim S, Misra A (2007) SNP genotyping: technologies and biomedical applications. Annu Rev Biomed Eng 9:289–320
Kim HJ, Baek KH, Lee SW, Kim JE, Lee BW, Cho HS, Kim WT, Choi D, Hur CG (2008) Pepper EST database: comprehensive in silico tool for analyzing the chili pepper (Capsicum annuum) transcriptome. BMC Plant Biol 8:101
Kim S, Park M, Yeom S, Kim Y-M, Lee JM, Lee H-A, Seo E, Choi J, Cheong K, Kim K-T, Jung K, Lee G-W, Oh S-G, Bae C-Y, Kim S-B, Lee H-Y, Kim S-Y, Kim M-S, Kang B-C, Jo YD, Yang H-B, Jeong H-J, Kang W-H, Kwon J-K, Shin C, Lim JY, Park JH, Huh JH, Kim J-S, Kim B-D, Cohen O, Paran I, Suh MC, Lee SB, Kim Y-K, Shin Y, Noh S-J, Park J, Seo YS, Kwon S-Y, Kim HA, Park JM, Kim H-J, Choi S-B, Bosland PW, Reeves G, Jo S-W, Lee B-W, Cho H-T, Choi H-S, Lee M-S, Yu Y, Choi YD, Park B-S, van Deynze A, Ashrafi H, Hill T, Kim WT, Pai H-S, Ahn HK, Yeam I, Giovannoni JJ, Rose JKC, Sørensen I, Lee S-J, Kim RW, Choi I-Y, Choi B-S, Lim J-S, Lee Y-H, Choi D (2014) Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nat Genet 46:270–278
Koo DH, Jo S-H, Bang J-W, Park H-M, Lee SH, Choi D (2008) Integration of cytogenetic and genetic linkage maps unveils the physical architecture of tomato chromosome 2. Genetics 179:1211–1220
Koumbaris GL, Bass HW (2003) A new single-locus cytogenetic mapping system for maize (Zea mays L.): overcoming FISH detection limits with marker-selected sorghum (S. propinquum L.) BAC clones. Plant J 35:647–659
Kwon J-K, Kim BD (2009) Localization of 5S and 25S rRNA genes on Somatic and meiotic chromosomes in Capsicum species of chili pepper. Mol Cells 27:205–209
Lanteri S, Pickersgill B (1993) Chromosomal structural-changes in Capsicum annuum L. and C. chinense Jacq. Euphytica 67:155–160
Lee JM, Nahm SH, Kim YM, Kim BD (2004) Characterization and molecular genetic mapping of microsatellite loci in pepper. Theor Appl Genet 108:619–627
Lee HR, Bae IH, Park SW, Kim HJ, Min WK, Han JH, Kim KT, Kim BD (2009) Construction of an Integrated Pepper Map Using RFLP, SSR, CAPS, AFLP, WRKY, rRAMP, and BAC End Sequences. Mol Cells 27:21–37
Lefebvre V, Pflieger S, Thabuis A, Caranta C, Blattes A, Chauvet JC, Daubeze AM, Palloix A (2002) Towards the saturation of the pepper linkage map by alignment of three intraspecific maps including known-function genes. Genome 45:839–854
Lehmensiek A, Sutherland MW, McNamara RB (2008) The use of high resolution melting HRM to map single nucleotide polymorphism markers linked to a covered smut resistance gene in barley. Theor Appl Genet 117:721–728
Livingstone KD, Lackney VK, Blauth JR, Van Wijk R, Jahn MK (1999) Genome mapping in Capsicum and the evolution of genome structure in the Solanaceae. Genetics 152:1183–1202
Lu FH, Kwon SW, Yoon MY, Kim KT, Cho MC, Yoon MK, Park YJ (2012) SNP marker integration and QTL analysis of 12 agronomic and morphological traits in F8 RILs of pepper (Capsicum annuum L.). Mol and Cells 34:25–34
Miller RT, Christoffels AG, Gopalakrishnan C, Burke J, Ptitsyn AA, Broveak TR, Hide WA (1999) A comprehensive approach to clustering of expressed human gene sequence: the sequence tag alignment and consensus knowledge base. Genome Res 9:1143–1155
Park YK, Kim BD, Kim BS, Armstrong KC, Kim NS (1999) Karyotyping of the chromosomes and physical mapping of the 5S rRNA and 18S-26S rRNA gene families in five different species in Capsicum. Genes Genet Syst 74:149–157
Park SW, An SJ, Yang HB, Kwon JK, Kang BC (2009) Optimization of high resolution melting analysis and discovery of single nucleotide polymorphism in Capsicum. Hort Environ Biotechnol 50:31–39
Pickersgill B (1971) Relationships between weedy and cultivated forms in some species of chilli peppers (genus Capsicum). Evolution 25:683–691
Piquemal J, Cinquin E, Couton F, Rondeau C, Seignoret E, Doucet I, Perret D, Villeger MJ, Vincourt P, Blanchard P (2005) Construction of an oilseed rape (Brassica napus L.) genetic map with SSR markers. Theor Appl Genet 111:1514–1523
Pochard E (1970) Description of trisomic individuals of Capsicum annuum L. obtained in progeny of a haploid plant. Ann Amel Plantes 20:233–256
Prince JP, Pochard E, Tanksley SD (1993) Construction of molecular linkage map of pepper and a comparison of synteny with tomato. Genome 36:404–417
Reed GH, Wittwer CT (2004) Sensitivity and specificity of single-nucleotide polymorphism scanning by high-resolution melting analysis. Clin Chem 50:1748–1754
Ririe KM, Rasmussen RP, Wittwer CT (1997) Product differentiation by analysis of DNA melting curves during the polymerase chain reaction. Anal Biochem 245:154–160
Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. Bioinformatics Methods and Protocols 132:365–386
Schiex T, Gaspin C (1997) CARTHAGENE: constructing and joining maximum likelihood genetic maps. Proc Int Conf Intell Syst Mol Biol 5:258–267
Shirasawa K, Asamizu E, Fukuoka H, Ohyama A, Sato S, Nakamura Y, Tabata S, Sasamoto S, Wada T, Kishida Y, Tsuruoka H, Fujishiro T, Yamada M, Isobe S (2010) An interspecific linkage map of SSR and intronic polymorphism markers in tomato. Theor Appl Genet 121:731–739
Sugita T, Semi Y, Sawada H, Utoyama Y, Hosomi Y, Yoshimoto E, Maehata Y, Fukuoka H, Nagata R, Ohyama A (2013) Development of simple sequence repeat markers and construction of a high-density linkage map of Capsicum annuum. Mol Breeding 31:909–920
Tanksley SD (1984) Linkage relationships and chromosomal locations of enzyme-coding genes in pepper, Capsicum annuum. Chromosoma 89:352–360
Tanksley SD, Bernatzky R, Lapitan NL, Prince JP (1988) Conservation of gene repertoire but not gene order in pepper and tomato. Proc Natl Acad Sci USA 85:6419–6423
Van Deynze A, Stoffel K, Buell CR, Kozik A, Liu J, Van der Knaap E, Francis D (2007) Diversity in conserved genes in tomato. BMC Genom 8:465
Wittwer CT, Reed GH, Gundry CN, Vandersteen JG, Pryor RJ (2003) High-resolution genotyping by amplicon melting analysis using LCGreen. Clin Chem 49:853–860
Wu F, Mueller LA, Crouzillat D, Petiard V, Tanksley SD (2006) Combining bioinformatics and phylogenetics to identify large sets of single-copy orthologous genes (COSII) for comparative, evolutionary and systematic studies: a test case in the euasterid plant clade. Genetics 174:1407–1420
Wu F, Eannetta NT, Xu Y, Durrett R, Mazourek M, Jahn MK, Tanksley SD (2009) A COSII genetic map of the pepper genome provides a detailed picture of synteny with tomato and new insights into recent chromosome evolution in the genus Capsicum. Theor Appl Genet 118:1279–1293
Yang W, Bai X, Kabelka E, Eaton C, Kamoun S, Van der Knaap E, Francis D (2004) Discovery of single nucleotide polymorphisms in Lycopersicon esculentum by computer aided analysis of expressed sequence tags. Mol Breeding 14:21–24
Acknowledgments
This work was supported by the grants from the Golden Seed Project (213002-04-1-CG910), Ministry of Agriculture, Food and Rural Affairs (MAFRA), Ministry of Oceans and Fisheries (MOF), Rural Development Administration (RDA) and Korea Forest Service (KFS), the Next-Generation BioGreen 21 Program (Plant Molecular Breeding Center No. PJ00906501), Rural Development Administration, and the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2012R1A1A3015216) Republic of Korea.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Park, SW., Jung, JK., Choi, EA. et al. An EST-based linkage map reveals chromosomal translocation in Capsicum . Mol Breeding 34, 963–975 (2014). https://doi.org/10.1007/s11032-014-0089-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11032-014-0089-0