Abstract
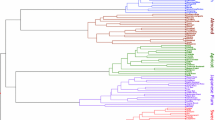
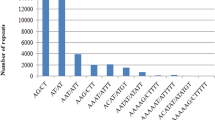
Ricinus communis is a versatile industrial oil crop that is cultivated worldwide. Genetic improvement and marker-assisted breeding of castor bean have been slowed owing to the lack of abundant and efficient molecular markers. As co-dominant markers, simple sequence repeats (SSRs) are useful for genetic evaluation and molecular breeding. The recently released whole-genome sequence of castor bean provides useful genomic resources for developing markers on a genome-wide scale. In the present study, the distribution and frequency of microsatellites in the castor bean genome were characterised and numerous SSR markers were developed using genomic data mining. In total, 18,647 SSR loci at a density of one SSR per 18.89 Kb in the castor bean genome sequence (representing approximately 352.27 Mb) were identified. Dinucleotide repeats were the most frequently observed microsatellites, although the AAT repeat motif was also prevalent. Using six cultivars as screening samples, 670 polymorphic SSR markers from 1,435 primer pairs (46.7 %) were developed. Trinucleotide motif loci contained a higher proportion of polymorphisms (48.5 %) than dinucleotide motif loci (39.2 %). The polymorphism level in the SSR loci was positively correlated with the increasing number of repeat units in the microsatellites. The phylogenetic relationship among 32 varieties was evaluated using the developed SSR markers. Cultivars developed at the same institute clustered together, suggesting that these cultivars have a narrow genetic background. The large number of SSR markers developed in this study will be useful for genetic mapping and for breeding improved castor-oil plants. These markers will also facilitate genetic and genomic studies of Euphorbiaceae.



Similar content being viewed by others
References
Allan G, Williams A, Rabinowicz PD, Chan AP, Ravel J, Keim P (2008) Worldwide genotyping of castor bean germplasm (Ricinus communis L.) using AFLPs and SSRs. Genet Resour Crop Evol 55:365–378
Anjani K (2012) Castor genetic resources: a primary gene pool for exploitation. Ind Crops Prod 35:1–14
Bajay MM, Pinheiro JB, Batista CEA, Nobrega MBD, Zucchi MI (2009) Development and characterization of microsatellite markers for castor (Ricinus communis L.), an important oleaginous species for biodiesel production. Conserv Genet Resour 1:237–239
Bajay MM, Zucchi MI, Kiihl TAM, Batista CEA, Monteiro M, Pinheiro JB (2011) Development of a novel set of microsatellite markers for Castor bean, Ricinus communis (Euphorbiaceae). Am J Bot 98:e87–e89
Caupin HJ (1997) Products from castor oil: past, present and future. In: Gunstone FD, Padley FB (eds) Lipid technologies and applications. Marcel Dekker, New York, pp 787–795
Cavagnaro PF, Senalik DA, Yang L, Simon PW, Harkins TT, Kodira CD, Huang S, Weng Y (2010) Genome-wide characterization of simple sequence repeats in cucumber (Cucumis sativus L.). BMC Genom 11:569
Chan AP, Crabtree J, Zhao Q, Lorenzi H, Orvis J, Puiu D, Melake-Berhan A, Jones KM, Redman J, Chen G, Cahoon EB, Gedil M, Stanke M, Haas BJ, Wortman JR, Fraser-Liggett CM, Ravel J, Rabinowicz PD (2010) Draft genome sequence of the oilseed species Ricinus communis. Nat Biotech 28:951–956
Chen YC, Liu T, Yu CH, Chiang TY, Hwang CC (2013) Effects of GC bias in next-generation-sequencing data on De Novo genome assembly. PLoS ONE 8(4):e62856. doi:10.1371/journal.pone.0062856
Comar V, Tilley D, Felix E, Turdera M, Neto MC (2004) Comparative energy evaluation of castor bean (Ricinus communis) production systems in Brazil and the U.S. In: Ortega E, Ulgiati S (eds) Proceedings of IV Biennial International Workshop ‘Advances in Energy Studies’. Unicamp, Campinas, pp 27–237
Eisen JA (1999) Mechanistic basis for microsatellite instability. In: Goldstein DB, Schlötterer C (eds) Microsatellites-evolution and application. Oxford University Press, Oxford
Foroni I, Woeste K, Monti LM, Rao R (2007) Identification of “Sorrento” walnut using simple sequence repeats (SSR’s). Genet Resour Crop Evol 54:1081–1094
Foster JT, Allan GJ, Chan AP, Rabinowicz PD, Ravel J, Jackson PJ, Keim P (2010) Single nucleotide polymorphisms for assessing genetic diversity in castor bean (Ricinus communis). BMC Plant Biol 10:3
Gajera BB, Kumar N, Singh AS, Punvar BS, Ravikiran R, Subhash N, Jadeja GC (2010) Assessment of genetic diversity in castor (Ricinus communis L.) using RAPD and ISSR markers. Ind Crop Prod 32:491–498
Govaerts R, Frodin DG, Radcliffe-Smith A (2000) World checklist and bibliography of Euphorbiaceae (with Pandaceae). Redwood Books Limited, Trowbridge
He ZH, Si EJ, Lai Y, Wang PX, Meng YX, Li BC, Ma XL, Shang XW, Wang HJ (2013) Genetic diversity and population structure in barley parent materials using SSR markers. J Triticeae Crops 33:894–900 (In Chinese)
Huo N, Lazo GR, Vogel JP, You FM, Ma Y, Hayden DM, Coleman-Derr D, Hill TA, Dvorak J, Anderson OD, Luo MC, Gu YQ (2008) The nuclear genome of Brachypodium distachyon: analysis of BAC end sequences. Funct Integr Genomics 8:135–147
IRGSP (International Rice Genome Sequencing Project) (2005) The map-based sequence of the rice genome. Nature 436:793–800
Jaillon O, Aury JM, Noel B, Policriti A, Clepet C et al (2007) The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449:463–467
Kalia RK, Rai MK, Kalia S, Singh R, Dhawan AK (2011) Microsatellite markers: an overview of the recent progress in plants. Euphytica 177:309–334
Kofler R, Schlotterer C, Lelley T (2007) SciRoKo: a new tool for whole genome microsatellite search and investigation. Bioinformatics 23:1683–1685
Li M, Yuyama N, Luo L, Hirata M, Cai HW (2009) In silico mapping of 1758 new SSR markers developed from public genomic sequences for sorghum. Mol Breed 24:41–47
Milani M, Dantas FV, Martins WFS (2009) Genetic divergence among castor bean genotypes by morphologic and molecular characters. Rev Bras Ol Fibros 13:61–71
Morgante M, Hanafey M, Powell W (2002a) Microsatellites are preferentially associated with nonrepetitive DNA in plant genomes. Nat Genet 30:194–200
Morgante M, Hanafey M, Powell W (2002b) Microsatellites are preferentially associated with nonrepetitive DNA in plant genomes. Nat Genet 30:194–200
Ogunniyi DS (2006) Castor oil: a vital industrial raw material. Bioresour Technol 97:1086–1091
Pranavi B, Sitaram G, Yamini KN, Kumar VD (2011) Development of EST-SSR markers in castor bean (Ricinus communis L.) and their utilization for genetic purity testing of hybrids. Genome 54:684–691
Price HJ, Dillon SL, Hodnett G, Rooney WL, Ross L et al (2005) Genome evolution in the genus Sorghum (Poaceae). Ann Bot 95:219–227
Qiu L, Yang C, Tian B, Yang JB, Liu A (2010) Exploiting EST databases for the development and characterization of EST-SSR markers in castor bean (Ricinus communis L.). BMC Plant Biol 10:278
Rivarola M, Foster JT, Chan AP, Williams AL, Rice D, Liu J, Melake-Berhan A, Hout Creasy H, Puiu D, Rosovitz MJ, Khouri HM, Beckstrom-Sternberg SM, Allan GJ, Keim P, Ravel J, Rabinowicz PD (2011) Organelle genome sequencing and worldwide genetic diversity of castor bean chloroplast genomes. PLoS ONE 6(7):e21743. doi:10.1371/journal.pone.0021743
Rohlf FJ (2000) NTSYSpc: numerical taxonomy and multivariate analysis system, Version 2.1, User Guide. Exeter Software, New York
Rozen S, Skaletsky HJ (2000) Primer3 on the WWW for general users and for biologist programmers. In: Krawetz SA, Misener S (eds) Bioinformatics methods and protocols: methods in molecular biology. Humana Press, Totowa/NJ, pp 365–386
Schlötterer C, Ritter R, Harr B, Brem G (1998) High mutation rate of a long microsatellite allele in Drosophila melanogaster provides evidence for allele-specific mutation rates. Mol Biol Evol 15:1269–1274
Scholz V, Silva JN (2008) Prospects and risks of the use of castor oil as a fuel. Biomass Bioenerg 32:95–100
Scott KD, Eggler P, Seaton G, Rossetto M, Ablett EM, Lee LS, Henry RJ (2000) Analysis of SSRs derived from grape ESTs. Theor Appl Genet 100:723–726
Seo KI, Lee GA, Ma KH, Hyun DY, Park YJ, Jung JW, Lee SY, Gwag JG, Kim CK, Lee MC (2011) Isolation and characterization of 28 polymorhpic SSR loci from castor bean (Ricinus communis L.). J Crop Sci Biotech 14:97–103
Sia EA, Jinks-Robertson S, Petes T (1997) Genetic control of microsatellite stability. Mutat Res 383:61–70
Tan ML, Jiang XH, Yan MF, Yan XC (2009) DNA extraction and establishment of SRAP reaction system in castor. Chin J Oil Crop Sci 31:531–536 (In Chinese)
Tan ML, Yan MF, Wang L, Wang LJ, Yan XC (2012) Analysis of genetic diversity in castor bean by SRAP marker. J Inner Mong Univ for Natly 27:533–540 (In Chinese)
Temnykh S, DeClerck G, Lukashova A, Lipovich L, Cartinhour S, McCouch S (2001) Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): frequency, length variation, transposon associations, and genetic marker potential. Genome Res 11:1441–1452
Thiel T, Michalek W, Varshney RK, Graner A (2003) Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theor Appl Genet 106:411–422
Tommasini L, Batley J, Arnold GM, Cooke RJ, Donini P, Lee D, Law JR, Lowe C, Moule C, Trick M, Edwards KJ (2003) The development of multiplex simple sequence repeat (SSR) markers to complement distinctness, uniformity and stability testing of rape (Brassica napus L.) varieties. Theor Appl Genet 106:1091–1101
Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S et al (2006) The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313:1596–1604
Ueno S, Moriguchi Y, Uchiyama K, Ujino-Ihara T, Futamura N, Sakurai T, Shinohara K, Tsumura Y (2012) A second generation framework for the analysis of microsatellites in expressed sequence tags and the development of EST-SSR markers for a conifer, Cryptomeria japonica. BMC Genom 13:136
Varshney RK, Graner A, Sorrells ME (2005a) Genic microsatellite markers in plants: features and applications. Trends Biotechnol 23:48–55
Varshney RK, Sigmund R, Börner A, Korzum V, Stein N, Sorrells ME, Langridge P, Graner A (2005b) Interspecific transferability and comparative mapping of barley EST-SSR markers in wheat, rye and rice. Plant Sci 168:195–202
Vasconcelos S, Onofre AVC, Milani M, Benko-Iseppon AM, Brasileiro-Vidal AC (2012) Molecular markers to access genetic diversity of castor bean: current status and prospects for breeding purposes. In: Ibrokhim A (ed) Plant breeding, ISBN: 978-953-307-932-5
Weber JL (1990) Informativeness of human (dC-dA)n.(dG-dT)n polymorphisms. Genomics 7:524–530
Wierdl M, Dominska M, Petes TD (1997) Microsatellite instability in yeast: dependence on the length of the microsatellite. Genetics 146:769–779
Yao QL, Yang KC, Pan GT, Rong TZ (2007) Genetic diversity of maize (Zea mays L.) landraces from Southwest China based on SSR data. J Genet Genomics 34:851–860
Yeh FC, Yang RC, Boyle T (1999) POPGENE, Microsoft Windowsbased freeware for population genetic analysis. Release 1.31. University of Alberta, Edmonton
Zhang R, Zhu AD, Wang JY, Zhang HR, Gao JS, Cheng YJ, Deng XX (2010) Development of Juglans Regia SSR markers by data mining of the EST database. Plant Mol Biol Rep 28:646–653
Zhao Y, Prakash CS, He G (2012) Characterization and compilation of polymorphic simple sequence repeat (SSR) markers of peanut from public database. BMC Res Notes 5:362
Zheng L, Qi JM, Fang PP, Su JG, Xu JT, Tao AF (2010) Genetic diversity and phylogenetic relationship of castor germplasm as revealed by SRAP analysis. J Wuhan Bot Res 28:1–6 (In Chinese)
Acknowledgments
We thank Dr Haibin Xu from the Institute of Medicinal Plant Development, Peking Union Medical College, for his assistance in SSR mining. We extend many thanks to anonymous reviewers for their constructive comments during the manuscript review. This work was jointly supported by the National Natural Science Foundation of China (Grant No. 31000726), the National Department (Agriculture) Public Benefit Scientific Research Foundation (Grant No. 201003057), and the Director Foundation of Oil Crops Research Institute of the Chinese Academy of Agricultural Sciences (Grant No. 1610172011009).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Tan, M., Wu, K., Wang, L. et al. Developing and characterising Ricinus communis SSR markers by data mining of whole-genome sequences. Mol Breeding 34, 893–904 (2014). https://doi.org/10.1007/s11032-014-0083-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11032-014-0083-6