Abstract
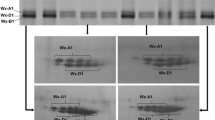
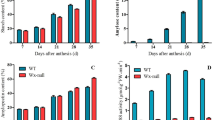
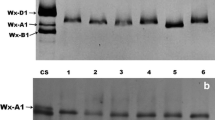
Waxy strains raised by waxy mutation have been found for three amaranth grains. Three genes encoding waxy protein were isolated from Amaranthus caudatus (Wx-ca), A. cruentus (Wx-cr), and A. hypochondriacus (Wx-hy). Sequence analysis indicated that the Wx-ca, Wx-cr, and Wx-hy genes contained the same exon (13 exons) and intron (12 introns) structure. The lengths of the Wx-ca, Wx-cr, and Wx-hy genes were 3,236, 3,237, and 3,225 bp, respectively. The alignment of the coding sequence of the three Waxy genes showed 12 polymorphic sites, including 11 single nucleotide polymorphisms (SNPs) (in exons 10 and 12 and introns 1, 3, 4, 9, and 11) and 5 deletions or insertions (indels) (in introns 4, 9, and 11). In particular, major polymorphism was detected in 8- and 3-bp indels in intron 4. Moreover, the mutation in the waxy alleles (wx-ca, wx-cr, and wx-hy) of all three species was also isolated and characterized. Comparison of coding sequences of the three Waxy genes and their waxy alleles indicated one base insertion (wx-ca: insert of T base in exon 8) and a base substitution (wx-cr: a G-to-T base substitution in exon 10; wx-hy: a G-to-A base substitution in exon 6), which occurred as internal termination codon in the three Waxy genes, suggesting the involvement of a nonsense or frameshift mutation. Therefore, these different mutations in coding regions were considered to be the cause of the waxy (amylose-free) phenotype.



Similar content being viewed by others
References
Ainsworth C, Gale M, Baird S (1983) The genetics of β-amylase isozymes in wheat. Theor Appl Genet 66:39–49
Arciga-Reyes L, Wootton L, Kieffer M, Davies B (2006) UPF1 is required for nonsense-mediated mRNA decay (NMD) and RNAi in arabidopsis. Plant J 47:480–489
Breathnach R, Chambon P (1981) Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem 50:349–383
Breene W (1991) Food uses of grain amaranth. Cereal Foods World 36:426–430
Bureau T, Wessler S (1992) Tourist: a large family of small inverted repeat elements frequently associated with maize genes. Plant Cell 4:1283–1294
Cai X, Wang Z, Xing Y, Zhang J, Hong M (1998) Aberrant splicing of intron 1 leads to the heterogeneous 5’UTR and decreased expression of waxy gene in rice cultivars of intermediate amylose content. Plant J 14:459–465
Camirand A, St-Pierre B, Marineau C, Brisson N (1990) Occurrence of a copia-like transposable element in one of the introns of the potato starch phosphorylase gene. Mol Gen Genet 224:33–39
Campbell WH, Gowri G (1990) Codon usage in higher plants, green algae, and cyanobacteria. Plant Physiol 92:1–11
Chan KF, Sun M (1997) Genetic diversity detected by isozyme and RAPD analysis of crop and wild species of amaranthus. Theor Appl Genet 95:865–873
Clark J, Robertson M, Ainsworth C (1991) Nucleotide sequence of a wheat (Triticum aestivum L.) cDNA clone encoding the waxy protein. Plant Mol Biol 16:1099–1101
Costea M, Sanders A, Waines G (2001) Preliminary results toward a revision of the Amaranthus hybridus species complex (Amaranthaceae). Sida 19:931–974
Costea M, Brenner DM, Tardif FJ, Tan YF, Sun M (2006) Delimitation of Amaranthus cruentus L. and Amaranthus caudatus L. using micromorphology and AFLP analysis: an application in germplasm identification. Genet Resour Crop Evol 53:1625–1633
Davis J, Supatcharee N, Khandellwal R, Chibbar R (2003) Synthesis of novel starches in Planta: opportunities and challenges. Starch/Staerke 55:107–120
Domon E, Fujita M, Ishikawa N (2002a) The insertion/deletion polymorphisms in the waxy gene of barley genetic resources from East Asia. Theor Appl Genet 104:132–138
Domon E, Saito A, Takeda K (2002b) Comparison of the waxy locus sequence from a non-waxy strain and two waxy mutants of spontaneous and artificial origins in barley. Genes Genet Syst 77:351–359
Dry I, Smith A, Edward A, Bhattacharyya M, Dunn P, Martin C (1992) Characterization of cDNAs encoding two isoforms of granule-bound starch synthase which show differential expression in developing storage organs of pea and potato. Plant J 2:193–202
Echt C, Schwartz D (1981) Evidence for the inclusion of controlling elements within the structural gene at the waxy locus in maize. Genetics 99:275–284
Emanuelsson O, Nielsen H, von Heijne G (1999) ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci 8:978
Fedoroff N, Wessler S, Shure M (1983) Isolation of the transposable maize controlling elements Ac and Ds. Cell 35:235–242
Fukunaga K, Kawase M, Kato K (2002) Structural variation in the Waxy gene and differentiation in foxtail millet [Setaria italica (L.) P. Beauv.]: implications for multiple origins of the waxy phenotype. Mol Genet Genomics 268:214–222
Furukawa K, Tagaya M, Inouye M, Preiss J, Fukui T (1990) Identification of lysine 15 at the active site in Escherichia coli glycogen synthase. Conservation of Lys-X-Gly-Gly sequence in the bacterial and mammalian enzymes. J Biol Chem 265:2086–2090
Hirano HY, Eiguchi M, Sano Y (1998) A single base change altered the regulation of the waxy gene at the posttranscriptional level during the domestication of rice. Mol Biol Evol 15:978–987
Hovenkamp-Hermelink J, Jacobsen E, Ponstein A, Visser R, Vos-Scheperkeuter G, Bijmolt E, Vries J, Witholt B, Feenstra W (1987) Isolation of an amylose-free starch mutant of the potato (Solanum tuberosum L.). Theor Appl Genet 75:217–221
Hseih J (1988) Genetic studies of the Wx gene of sorghum (Sorghum bicolor [L.] Moench). Bot Bull Academia Sinica 29:293–299
Hsieh J, Liu C, Hsing Y (1996) Molecular cloning of a sorghum cDNA encoding the seed waxy protein. Plant Physiol 112:1735
Inouchi N, Nishi K, Tanaka S, Asai M, Kawase Y, Hata Y, Konishi Y, Yue S, Fuwa H (1999) Characterization of amaranth and quinoa starches. J Appl Glycosci 46:233–240
Isshiki M, Yamamoto Y, Satoh H, Shimamoto K (2001) Nonsense-mediated decay of mutant waxy mRNA in rice. Plant Physiol 125:1388–1395
Juan R, Pastor J, Alaiz M, Vioque J (2007) Electrophoretic characterization of Amaranthus L. seed proteins and its systematic implications. Bot J Linn Soc 155:57–63
Kawase M, Fukunaga K, Kato K (2005) Diverse origins of waxy foxtail millet crops in East and Southeast Asia mediated by multiple transposable element insertions. Mol Genet Genomics 274:131–140
Kimura M (1980) A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Kimura T, Ideta O, Saito A (2000) Identification of the gene encoding granule-bound starch synthase I in sweet potato (Ipomoea batatas (L.) Lam.). Plant Biotechnol 17:247–252
Klosgen R, Gierl A, Schwarz-Sommer Z, Saedler H (1986) Molecular analysis of the waxy locus of Zea mays. Mol Genet Genomics 203:237–244
Konishi Y, Nojima H, Okuno K, Asaoka M, Fuwa H (1985) Characterization of starch granules from waxy, nonwaxy, and hybrid seeds of Amaranthus hypochondriacus L. Agri Biol Chem 49:1965–1971
Kumar A, Larsen C, Preiss J (1986) Biosynthesis of bacterial glycogen. Primary structure of Escherichia coli ADP-glucose: alpha-1, 4-glucan, 4-glucosyltransferase as deduced from the nucleotide sequence of the glgA gene. J Biol Chem 261:16256–16259
Lanoue KZ, Wolf PG, Browning S, Hood EE (1996) Phylogenetic analysis of restriction-site variation in wild and cultivated Amaranthus species (Amaranthaceae). Theor Appl Genet 93:722–732
Marcone M (2001) Starch properties of Amaranthus pumilus (seabeach amaranth): a threatened plant species with potential benefits for the breeding/amelioration of present Amaranthus cultivars. Food Chem 73:61–66
Mochizuki K, Umeda M, Ohtsubo H, Ohtsubo E (1992) Characterization of a plant SINE, p-SINE1, in rice genomes. Jpn J Genet 67:155–166
Murai J, Taira T, Ohta D (1999) Isolation and characterization of the three waxy genes encoding the granule-bound starch synthase in hexaploid wheat. Gene 234:71–79
Murray M, Thompson W (1980) Rapid isolation of high molecular weight plant DNA. Nucl Acids Res 8:4321–4325
Nakamura T, Vrinten P, Hayakawa K, Ikeda J (1998) Characterization of a granule-bound starch synthase isoform found in the pericarp of wheat. Plant Physiol 118:451–459
Nakayama H, Afzal M, Okuno K (1998) Intraspecific differentiation and geographical distribution of Wx alleles for low amylose content in endosperm of foxtail millet, Setaria italica (L.) Beauv. Euphytica 102:289–293
Nelson O, Rines H (1962) The enzymatic deficiency in the waxy mutant of maize. Biochem Biophys Res Commun 9:297–300
Okagaki R (1992) Nucleotide sequence of a long cDNA from the rice waxy gene. Plant Mol Biol 19:513–516
Okuno K, Sakaguchi S (1981) Glutinous and non-glutinous starches in perisperm of grain amaranths. Cereal Res Commun 9:305–310
Okuno K, Sakaguchi S (1982) Inheritance of starch characteristics in perisperm of Amaranthus hypochondriacus. J Hered 73:467
Park YJ, Nemoto K, Nishikawa T, Matsushima K, Minami M, Kawase M (2009) Molecular cloning and characterization of granule bound starch synthase I cDNA from a grain amaranth (Amaranthus cruentus L.). Breed Sci (in press)
Rohde W, Becker D, Salamini F (1988) Structural analysis of the waxy locus from Hordeum vulgare. Nucl Acids Res 16:7185
Salehuzzaman S, Jacobsen E, Visser R (1993) Isolation and characterization of a cDNA encoding granule-bound starch synthase in cassava (Manihot esculenta Crantz) and its antisense expression in potato. Plant Mol Biol 23:947–962
Sano Y (1984) Differential regulation of waxy gene expression in rice endosperm. Theor Appl Genet 68:467–473
Sauer JD (1950) The grain amaranths: a survey of their history and classification. Ann Missouri Bot Gard 37:561–619
Sauer JD (1967) The grain amaranths and their relatives: a revised taxonomic and geographic survey. Ann Missouri Bot Gard 54:103–137
Saunders R, Becker R (1984) Amaranthus: a potential food and feed resource. Adv Cereal Sci Technol 6:357–396
Sprague G, Brimhall B, Hixon R (1943) Some effects of the waxy gene in corn on properties of the endosperm starch. J Am Soc Agron 35:817
Sugimoto Y, Yamada K, Sakamoto S, Fuwa H (1981) Some properties of normal-and waxy-type starches of Amaranthus hypochondriacus L. pp 112–116
Swofford D (1988) PAUP*: phylogenetic analysis using parsimony and other methods, version 4.0 (test ver. 61–64). Sinauer Associates Publishers, Sunderland
Thompson JDG, Plewniak FJ, Higgins DG (1997) The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 24:4876–4882
Tsai C (1974) The function of the waxy locus in starch synthesis in maize endosperm. Biochem Genetics 11:83–96
van der Leij F, Visser R, Ponstein A, Jacobsen E, Feenstra W (1991) Sequence of the structural gene for granule-bound starch synthase of potato (Solanum tuberosum L.) and evidence for a single point deletion in the amf allele. Mol Gen Genet 228:240–248
Van K, Onoda S, Kim MY, Kim KD, Lee SH (2008) Allelic variation of the Waxy gene in foxtail millet [Setaria italica (L.) P. Beauv.] by single nucleotide polymorphisms. Mol Genet Genom 279:255–266
Vrinten P, Nakamura T, Yamamori M (1999) Molecular characterization of waxy mutations in wheat. Mol Genet Genomics 261:463–471
Wang Z, Wu Z, Xing Y, Zheng F, Guo X, Zhang W, Hong M (1990) Nucleotide sequence of rice waxy gene. Nucl Acids Res 18:5898
Wang Z, Zheng F, Shen G, Gao J, Snustad D, Li M, Zhang J, Hong M (1995) The amylose content in rice endosperm is related to the post-transcriptional regulation of the waxy gene. Plant J 7:613–622
Wessler S, Varagona M (1985) Molecular basis of mutations at the waxy locus of maize: correlation with the fine structure genetic map. Proc Natl Acad Sci 82:4177–4181
Xu F, Sun M (2001) Comparative analysis of phylogenetic relationships of grain amaranths and their wild relatives (Amaranthus; Amaranthaceae) using internal transcribedspacer, amplified fragment length polymorphism, and double-primer fluorescent intersimple sequence repeat markers. Mol Phylogenet Evol 21:372–387
Acknowledgments
We express our sincere thanks to Dr. D. Brenner of USDA-ARS-MWA-PIRU; Iowa State University, USA, for providing the accessions used in this experiment.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Park, YJ., Nemoto, K., Nishikawa, T. et al. Waxy strains of three amaranth grains raised by different mutations in the coding region. Mol Breeding 25, 623–635 (2010). https://doi.org/10.1007/s11032-009-9360-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11032-009-9360-1