Abstract
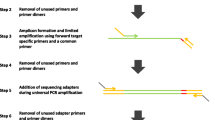
Mis-priming associated with uncharacterised single nucleotide polymorphisms (SNPs) may lead to failure of PCR for genotyping. This is particularly troublesome in high-throughput SNP genotyping applications relying on multiplex PCR (2–40-plex) generating many short amplicons (80–120 bp) of similar size, an approach best suited for whole genome scans. However, if the target SNPs are clustered within a few target genes one option to ameliorate this is to increase the amplicon length, effectively reducing the potential for primer/template interactions and mis-priming. We tested this approach in a diverse population of 372 Eucalyptus pilularis individuals (π = 8.11 × 10−3, H e = 0.75) using a modified Sequenom iPLEX gold assay. Four candidate genes (MYB1, MYB2, CAD and CCR) were amplified in a single long range multiplex capture PCR generating 6 long amplicons ranging in size from 907 to 2,225 bp. This contrasts with the standard approach which would have required the amplification of 98 short amplicons in 4 multiplex reactions. These 6 long amplicons provided the assay template for 98 assays (87 SNP and 11 InDel) within the 4 candidate genes. Reaction results indicated that longer amplicons could provide a suitable template for genotyping assays, with 90.8% of assays functional and 84.3% of assays suitable for downstream analysis. Additional advantages of this approach were the capacity for troubleshooting using gel electrophoresis and savings of 94% in capture primer synthesis costs. This approach will have the greatest relevance for candidate gene approaches for association testing in uncharacterised populations of organisms with high sequence diversity.



Similar content being viewed by others
References
Buetow KH, Edmonson M, MacDonald R, Clifford R, Yip P, Kelley J, Little DP, Strausberg R, Koester H, Cantor CR, Braun A (2001) High-throughput development and characterization of a genomewide collection of gene-based single nucleotide polymorphism markers by chip-based matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Proc Natl Acad Sci USA 98:581–584
Desai C, Madamwar D (2007) Extraction of inhibitor-free metagenomic DNA from polluted sediments, compatible with molecular diversity analysis using adsorption and ion-exchange treatments. Bioresour Technol 98:761–768
Devey M, Bell JC, Smith DN, Neale DB, Moran GF (1996) A genetic linkage map for Pinus radiata based on RFLP, RAPD, and microsatellite markers. Theor Appl Genet 92:673–679
Galvin P (2002) A nanobiotechnology roadmap for high-throughput single nucleotide polymorphism analysis. Psychiatr Genet 12:75–82
Goicoechea M, Lacombe E, Legay S, Mihaljevic S, Rech P, Jauneau A, Lapierre C, Pollet B, Verhaegen D, Chaubet-Gigot N, Grima-Pettenati J (2005) EgMYB2, a new transcriptional activator from Eucalyptus xylem, regulates secondary cell wall formation and lignin biosynthesis. Plant J 43:553–567
Gonzales-Martinez SC, Wheeler NC, Ersoz E, Nelson CD, Neale DN (2007) Association genetics in Pinus taeda L. I. Wood property traits. Genetics 175:399–409
Grima-Pettenati J, Feuillet C, Goffner D, Borderies G, Boudet AM (1993) Molecular cloning and expression of a Eucalyptus gunnii cDNA clone encoding cinnamyl alcohol dehydrogenase. Plant Mol Biol 21:1085–1095
Henegariu O, Heerema NA, Dlouhy SR, Vance GH, Vogt PH (1997) Multiplex PCR: critical parameters and step-by-step protocol. Biotechniques 23:504–511
Ingvarsson PK, Garcia MV, Luquez V, Hall D, Jansson S (2008) Nucleotide polymorphism and phenotypic associations within and around the phytochrome B2 locus in European aspen (Populus tremula, Salicaceae). Genetics 178:2217–2226
Kim CS, Lee CH, Shin JS, Chung YS, Hyung NI (1997) A simple and rapid method for isolation of high quality genomic DNA from fruit trees and conifers using PVP. Nucleic Acids Res 25:1085–1086
Konieczny A, Ausubel FM (1993) A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J 4:403–410
Legay S, Lacombe E, Goicoechea M, Brière C, Séguin A, Mackay J, Grima-Pettenati J (2007) Molecular characterization of EgMYB1, a putative transcriptional repressor of the lignin biosynthetic pathway. Plant Sci 173:542–549
Li P, Kupfer KC, Davies CJ, Burbee D, Evans GA, Garner HR (1997) PRIMO: a primer design program that applies base quality statistics for automated large-scale DNA sequencing. Genomics 40:476–485
Masouleh AK, Waters DL, Reinke RF, Henry RJ (2009) A high-throughput assay for rapid and simultaneous analysis of perfect markers for important quality and agronomic traits in rice using multiplexed MALDI-TOF mass spectrometry. Plant Biotechnol J 7:355–363
Oeth P, Beaulieu M, Park C, Kosman D, del Mistro G, van den Boom D, Jurinke C (2008) iPLEX gold assay for SNP genotyping biotechniques. Protocol Guide 2008:37
Pattemore JA, Trau M, Henry RJ (2007) Nanotechnology: the future of cost-effective plant genotyping. In: Henry RJ (ed) Plant genotyping II. CABI, Wallingford, pp 133–153
Poke FS, Vaillancourt RE, Elliott RC, Reid JB (2003) Sequence variation in two lignin biosynthesis genes, cinnamoyl CoA reductase (CCR) and cinnamyl alcohol dehydrogenase 2 (CAD2). Mol Breeding 12:107–118
Ragoussis J, Elvidge GP, Kaur K, Colella S (2006) Matrix-assisted laser desorption/ionisation, time-of-flight mass spectrometry in genomics research. PLoS genetics 2:e100
Ross P, Hall L, Smirnov I, Haff L (1998) High level multiplex genotyping by MALDI-TOF mass spectrometry. Nat Biotechnol 16:1347–1351
Sachidanandam R, Weissman D, Schmidt SC, Kakol JM, Stein LD, Marth G, Sherry S, Mullikin JC, Mortimore BJ, Willey DL, Hunt SE, Cole CG, Coggill PC, Rice CM, Ning Z, Rogers J, Bentley DR, Kwok PY, Mardis ER, Yeh RT, Schultz B, Cook L, Davenport R, Dante M, Fulton L, Hillier L, Waterston RH, McPherson JD, Gilman B, Schaffner S, Van Etten WJ, Reich D, Higgins J, Daly MJ, Blumenstiel B, Baldwin J, Stange-Thomann N, Zody MC, Linton L, Lander ES, Altshuler D (2001) A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature 409:928–933
Shepherd M, Cross M, Stokoe R, Scott L, Jones M (2002) High-throughput DNA extraction from forest trees. Plant Mol Biol Rep 20:425
Shepherd M, Sexton TR, Thomas D, Henson M (2009) Utility of symphyomyrt-derived microsatellite markers (Genus Eucalyptus Subgenus Symphomyrtus) in Eucalyptus pilularis (Subgenus Eucalyptus) (submitted)
Sladek R, Rocheleau G, Rung J, Dina C, Shen L, Serre D, Boutin P, Vincent D, Belisle A, Hadjadj S, Balkau B, Heude B, Charpentier G, Hudson TJ, Montpetit A, Pshezhetsky AV, Prentki M, Posner BI, Balding DJ, Meyre D, Polychronakos C, Froguel P (2007) A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature 445:881–885
Smith LM (1993) The future of DNA sequencing. Science 262:530–532
Stewart NC, Via LE (1993) A rapid CTAB DNA isolation technique useful for RAPD fingerprinting and other PCR applications. Biotechniques 14:748–749
Stoeckel S, Grange J, Fernandez-Manjarres JF, Bilger I, Frascaria-Lacoste N, Mariette S (2006) Heterozygote excess in a self-incompatible and partially clonal forest tree species Prunus avium L. Mol Ecol 15:2109–2118
Thumma BR, Nolan MF, Evans R, Moran GF (2005) Polymorphisms in cinnamoyl CoA reductase (CCR) are associated with variation in microfibril angle in Eucalyptus spp. Genetics 171:1257–1265
Tibbits JFG, McManus LJ, Spokevicius AV, Bossinger G (2006) A rapid method for tissue collection and high throughput genomic DNA isolation from mature trees. Plant Mol Biol Rep 24:81–91
Ward M, Dick CW, Gribel R, Lowe AJ (2005) To self, or not to self…a review of outcrossing and pollen-mediated gene flow in neotropical trees. Heredity 95:246–254
Zhao Z, Fu YX, Hewett-Emmett D, Boerwinkle E (2003) Investigating single nucleotide polymorphism (SNP) density in the human genome and its implications for molecular evolution. Gene 312:207–213
Acknowledgments
This research was funded by the CRC for Forestry. The authors thank Forests NSW for providing leaf samples, as well as Chris Harwood, Peter Bundock, Julie Pattemore, Helen Hill, Dan Waters and Ardashir Kharabian Masouleh for many helpful comments and suggestions.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sexton, T.R., Henry, R.J., McManus, L.J. et al. Capture of assay template by multiplex PCR of long amplicons for genotyping SNPs and InDels with MALDI-TOF mass spectrometry. Mol Breeding 25, 471–480 (2010). https://doi.org/10.1007/s11032-009-9345-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11032-009-9345-0