Abstract
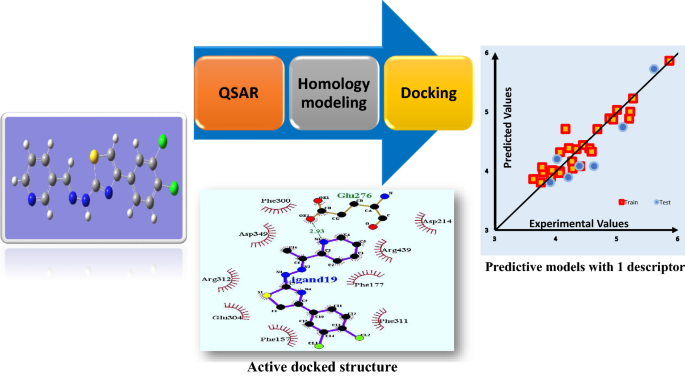
Quantitative structure–activity relationships (QSAR) and molecular docking studies have been performed on a series of 35 α-glucosidase inhibitory derivatives. The QSAR models have been developed by genetic algorithm-multiple linear regression (GA-MLR) and least squares-support vector machine (LS-SVM) methods to correlate the conformational descriptors to the inhibitory activity. The obtained models with 5 descriptors were validated and illustrated to be statistically significant. They had desirable prediction based on squared correlation coefficient (R2), cross-validated correlation coefficient (Q2), root-mean-squares error (RMSE) and Fisher (F) parameters (R2 = 0.951, Q2 = 0.931, RMSE = 0.121, and F = 114.629 for GA-MLR model, and R2 = 0.989, Q2 = 0.987, RMSE = 0.056 and F = 543.754 for LS-SVM model). The crucial descriptor named DELS was explored to have the highest correlation with the inhibitory activity and thus has been chosen to build a simple model. The QSAR model developed with this mono-descriptor showed appropriate results of the predicted model using LS-SVM method (R2 = 0.888, Q2 = 0.872, RMSE = 0.185 and F = 221.459). Also, molecular docking which focuses on the interaction between ligands and α-glucosidase in the protein active site considered different binding positions to find the best binding mode. It helped the QSAR study to propose more comprehensive details of the compounds structures and was used to design more active compounds. The most active designed compound had a high inhibitory activity of 9.22 that can be proposed for the treatment of diabetes type 2.
Graphic abstract

















Similar content being viewed by others
References
Imran S, Taha M, Ismail NH, Kashif SM, Rahim F, Jamil W, Hariono M, Yusuf M, Wahab H (2015) Synthesis of novel flavone hydrazones: in-vitro evaluation of α-glucosidase inhibition, QSAR analysis and docking studies. Eur J Med Chem 105:156–170. https://doi.org/10.1016/j.ejmech.2015.10.017
Goldenberg RM (2011) Management of unmet needs in type 2 diabetes mellitus: the role of incretin agents. Can J Diabetes 35(5):518–527. https://doi.org/10.1016/S1499-2671(11)80008-0
Narender T, Madhur G, Jaiswal N, Agrawal M, Maurya CK, Rahuja N, Srivastava AK, Tamrakar AK (2013) Synthesis of novel triterpene and N-allylated/N-alkylated niacin hybrids as α-glucosidase inhibitors. Eur J Med Chem 63:162–169. https://doi.org/10.1016/j.ejmech.2013.01.053
Dinparast L, Valizadeh H, Bahadori MB, Soltani S, Asghari B, Rashidi MR (2016) Design, synthesis, α-glucosidase inhibitory activity, molecular docking and QSAR studies of benzimidazole derivatives. J Mol Struct 1114:84–94. https://doi.org/10.1016/j.molstruc.2016.02.005
Park H, Hwang KY, Kim YH, Oh KH, Lee JY, Kim K (2008) Discovery and biological evaluation of novel α-glucosidase inhibitors with in vivo antidiabetic effect. Bioorg Med Chem Lett 18(13):3711–3715. https://doi.org/10.1016/j.bmcl.2008.05.056
Park H, Hwang KY, Oh KH, Kim YH, Lee JY, Kim K (2008) Discovery of novel α-glucosidase inhibitors based on the virtual screening with the homology-modeled protein structure. Bioorg Med Chem 16(1):284–292. https://doi.org/10.1016/j.bmc.2007.09.036
Asadollahi-Baboli M, Dehnavi S (2018) Docking and QSAR analysis of tetracyclic oxindole derivatives as α-glucosidase inhibitors. Comput Biol Chem 76:283–292. https://doi.org/10.1016/j.compbiolchem.2018.07.019
Scott LJ, Spencer CM (2000) Miglitol: a review of its therapeutic potential in type 2 diabetes. Drugs 59(3):521–549. https://doi.org/10.2165/00003495-200059030-00012
Wang SL (2018) New novel α-glucosidase inhibitors produced by microbial conversion. Process Biochem 65:228–232. https://doi.org/10.1016/j.procbio.2017.11.016
Channar PA, Saeed A, Larik FA, Rashid S, Iqbal Q, Rozi M, Younis S, Mahar J (2017) Design and synthesis of 2, 6-di (substituted phenyl) thiazolo [3, 2-b]-1, 2, 4-triazoles as α-glucosidase and α-amylase inhibitors, co-relative pharmacokinetics and 3D QSAR and risk analysis. Biomed Pharmacother 94:499–513. https://doi.org/10.1016/j.biopha.2017.07.139
Ali F, Khan KM, Salar U, Taha M, Ismail NH, Wadood A, Riaz M, Perveen S (2017) Hydrazinyl arylthiazole based pyridine scaffolds: synthesis, structural characterization, in vitro α-glucosidase inhibitory activity, and in silico studies. Eur J Med Chem 138:255–272. https://doi.org/10.1016/j.ejmech.2017.06.041
Ghaslani D, Gorji ZE, Gorji AE, Riahi S (2017) Descriptive and predictive models for Henry’s law constant of CO2 in ionic liquids: a QSPR study. Chem Eng Res Des 120:15–25. https://doi.org/10.1016/j.cherd.2016.12.020
Hasanebrahimi G, Riahi S, Fini MF (2017) Exploring beneficial structural features of ionic surfactants for wettability alteration of carbonate rocks using QSPR modeling technique. J Mol Liq 240:196–208. https://doi.org/10.1016/j.molliq.2017.05.009
Mehraein I, Riahi S (2017) The QSPR models to predict the solubility of CO2 in ionic liquids based on least-squares support vector machines and genetic algorithm-multi linear regression. J Mol Liq 225:521–530. https://doi.org/10.1016/j.molliq.2016.10.133
Abbasi-Radmoghaddam Z, Riahi S, Gharaghani S, Mohammadi-Khanaposhtanai M (2020) Design of potential anti-tumor PARP-1 inhibitors by QSAR and molecular modeling studies. Mol Divers. https://doi.org/10.1007/s11030-020-10063-9
Rezaei B, Riahi S (2016) Prediction of CO2 loading of amines in carbon capture process using membrane contactors: a molecular modeling. J Nat Gas Sci Eng 33:388–396. https://doi.org/10.1016/j.jngse.2016.05.003
Liu Z, Liu Y, Zeng G, Shao B, Chen M, Li Z, Jiang Y, Liu Y, Zhang Y, Zhong H (2018) Application of molecular docking for the degradation of organic pollutants in the environmental remediation: a review. Chemosphere 203:139–150. https://doi.org/10.1016/j.chemosphere.2018.03.179
Liu Y, Ke Z, Cui J, Chen WH, Ma L, Wang B (2008) Synthesis, inhibitory activities, and QSAR study of xanthone derivatives as α-glucosidase inhibitors. Bioorg Med Chem 16(15):7185–7192. https://doi.org/10.1016/j.bmc.2008.06.043
Kraim K, Khatmi D, Saihi Y, Ferkous F, Brahimi M (2009) Quantitative structure activity relationship for the computational prediction of α-glucosidase inhibitory. Chemom Intell Lab Syst 97(2):118–126. https://doi.org/10.1016/j.chemolab.2009.03.006
Release H (2002) 7.5 for windows, molecular modeling system, Hypercube. Inc. http://www.hyper.com
Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Scalmani G, Barone V, Mennucci B, Petersson GA, Nakatsuji H (2009) Gaussian 09, Gaussian, Inc., Wallingford, CT, vol 32, pp 5648–5652
Todeschini R, Consonni V, Mauri A, Pavan M (2002) DRAGON software. Milano, Italy
Gagic Z, Nikolic K, Ivkovic B, Filipic S, Agbaba D (2016) QSAR studies and design of new analogs of vitamin E with enhanced antiproliferative activity on MCF-7 breast cancer cells. J Taiwan Inst Chem Eng 59:33–44. https://doi.org/10.1016/j.jtice.2015.07.019
Roy K, Kar S, Das RN (2015) Understanding the basics of QSAR for applications in pharmaceutical sciences and risk assessment. Academic Press, London
Golbraikh A, Tropsha A (2000) Predictive QSAR modeling based on diversity sampling of experimental datasets for the training and test set selection. Mol Divers 5(4):231–243. https://doi.org/10.1023/A:1021372108686
Hawkins DM, Basak SC, Mills D (2003) Assessing model fit by cross-validation. J Chem Inf Comput Sci 43(2):579–586. https://doi.org/10.1021/ci025626i
Gharaghani S, Khayamian T, Ebrahimi M (2013) Molecular dynamics simulation study and molecular docking descriptors in structure-based QSAR on acetylcholinesterase (AChE) inhibitors. SAR QSAR Environ Res 24(9):773–794. https://doi.org/10.1080/1062936X.2013.792877
Aouidate A, Ghaleb A, Ghamali M, Chtita S, Choukrad M, Sbai A, Bouachrine M, Lakhlifi T (2016) Combining DFT and QSAR studies for predicting psychotomimetic activity of substituted phenethylamines using statistical methods. J Taibah Univers Sci 10(6):787–796. https://doi.org/10.1016/j.jtusci.2016.07.001
Pitman MR, Menz RI (2006). Methods for protein homology modelling. In: Applied mycology and biotechnology, vol 6. Elsevier, pp 37–59
https://www.uniprot.org/uniprot/P53341.fasta. Accessed Sept 2018
Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31(2):455–461. https://doi.org/10.1016/S1874-5334(06)80005-5
Laskowski RA, Swindells MB. (2011). LigPlot+: multiple ligand–protein interaction diagrams for drug discovery, pp 2778–2786. https://doi.org/10.1021/ci200227u
He J, Peng T, Yang X, Liu H (2018) Development of QSAR models for predicting the binding affinity of endocrine disrupting chemicals to eight fish estrogen receptor. Ecotoxicol Environ Saf 148:211–219. https://doi.org/10.1016/j.ecoenv.2017.10.023
Jouyban A, Shayanfar A, Ghafourian T, Acree WE Jr (2014) Solubility prediction of pharmaceuticals in dioxane + water mixtures at various temperatures: effects of different descriptors and feature selection methods. J Mol Liq 195:125–131. https://doi.org/10.1016/j.molliq.2014.02.012
Jukić M, Rastija V, Opačak-Bernardi T, Stolić I, Krstulović L, Bajić M, Glavaš-Obrovac L (2017) Antitumor activity of 3, 4-ethylenedioxythiophene derivatives and quantitative structure-activity relationship analysis. J Mol Struct 1133:66–73. https://doi.org/10.1016/j.molstruc.2016.11.074
Roy K, Das RN (2013) QSTR with extended topochemical atom (ETA) indices. 16. Development of predictive classification and regression models for toxicity of ionic liquids towards Daphnia magna. J Hazard Mater 254:166–178. https://doi.org/10.1016/j.jhazmat.2013.03.023
Durrant JD, McCammon JA (2011) BINANA: a novel algorithm for ligand-binding characterization. J Mol Gr Model 29(6):888–893. https://doi.org/10.1016/j.jmgm.2011.01.004
Acknowledgments
The authors would like to gratefully acknowledge the support from the Institute of Petroleum Engineering (IPE), University of Tehran.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the supplementary material.
Rights and permissions
About this article
Cite this article
Izadpanah, E., Riahi, S., Abbasi-Radmoghaddam, Z. et al. A simple and robust model to predict the inhibitory activity of α-glucosidase inhibitors through combined QSAR modeling and molecular docking techniques. Mol Divers 25, 1811–1825 (2021). https://doi.org/10.1007/s11030-020-10164-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11030-020-10164-5