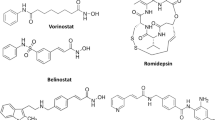
Abstract
As one of the hot topics in the epigenetic studies, histone deacetylases inhibitors (HDACIs) have been introduced to treat a variety of diseases such as cancer, immune disorder and neuronal diseases. Given the high numbers of available pathways in which HDACs are involved, the HDACIs that act particularly on Class I or Class II enzymes are considered as possible candidates for anticancer drugs. Due to their effective roles in the onset of cancer and its progression, HDAC Class I isoforms (HDAC 1, 2, 3 and 8) were considered in this study. Herein, our objective is to determine the important isoform-selective and isoform-active structural features of HDACIs using the valid classification models. For this purpose, a diverse dataset comprising 8224 HDAC modulators was collected from the binding database. To identify the significant discriminative features, five classification models were generated by supervised Kohonen network and support vector machine methods. Variable importance in projection method was used as a variable selection approach. The results obtained from descriptor analysis show that physicochemical properties, such as hydrogen bonding, number of branches, size, flexibility, polarity and sphericity in the structure of molecules, were closely related to the bioactivity of HDACIs. The reliability and predictive ability of the conducted models were evaluated using the tenfold cross-validation techniques, test sets and applicability domain analysis. All of the obtained classification models represented high statistical quality and predictive ability with accuracy greater than 85% for the test sets. The proposed strategy and the selective patterns represented in this paper can be applied by researchers in the pharmaceutical sciences who aim to use the same idea for the design of drugs with improved anticancer properties.
Graphic abstract







Similar content being viewed by others
References
Thangapandian S, John S, Lee KW (2012) Molecular dynamics simulation study explaining inhibitor selectivity in different class of histone deacetylases. J Biomol Struct Dyn 29(4):677–698. https://doi.org/10.1080/07391102.2012.10507409
Bolden JE, Shi W, Jankowski K, Kan CY, Cluse L, Martin BP, MacKenzie KL, Smyth GK, Johnstone RW (2013) HDAC inhibitors induce tumor-cell-selective pro-apoptotic transcriptional responses. Cell Death Dis 4(2):519–534. https://doi.org/10.1038/cddis.2013.9
Gao S, Zang J, Gao Q, Liang X, Ding Q, Li X, Xu W, Chou CJ, Zhang Y (2017) Design, synthesis and anti-tumor activity study of novel histone deacetylase inhibitors containing isatin-based caps and o-phenylenediamine-based zinc binding groups. Bioorg Med Chem 25(12):2981–2994. https://doi.org/10.1016/j.bmc.2017.03.036
Zhang J, Zhong Q (2014) Histone deacetylase inhibitors and cell death. Cell Mol Life Sci 71:3885–3901. https://doi.org/10.1007/s00018-014-1656-6
Falkenberg KJ, Johnstone RW (2014) Histone deacetylases and their inhibitors in cancer, neurological diseases and immune disorders. Nat Rev Drug Discov 13:673–691. https://doi.org/10.1038/nrd4360
Haberland M, Montgomery RL, Olson EN (2009) The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat Rev Genet 10:32–42. https://doi.org/10.1038/nrg2485
Yoon S, Eom GH (2016) HDAC and HDAC inhibitor: from cancer to cardiovascular diseases. Chonnam Med J 52(1):1–11. https://doi.org/10.4068/cmj.2016.52.1.1
Li Y, Seto E (2016) HDACs and HDAC inhibitors in cancer development and therapy. Cold Spring Harbor Perspect Med 6(10):26831–26872. https://doi.org/10.1101/cshperspect.a026831
Barneda-Zahonero B, Parra M (2012) Histone deacetylases and cancer. Mol Oncol 6(6):579–589. https://doi.org/10.1016/j.molonc.2012.07.003
Mottamal M, Zheng S, Huang TL, Wang G (2015) Histone deacetylase inhibitors in clinical studies as templates for new anticancer agents. Molecules 20(3):3898–3941. https://doi.org/10.3390/molecules20033898
Mai A, Massa S, Rotili D, Cerbara I, Valente S, Pezzi R, Simeoni S, Ragno R (2005) Histone deacetylation in epigenetics: an attractive target for anticancer therapy. Med Res Rev 25(3):261–309. https://doi.org/10.1002/med.20024
Goey AK, Sissung TM, Peer CJ, Figg WD (2016) Pharmacogenomics and histone deacetylase inhibitors. Pharmacogenom J 16:1807–1815. https://doi.org/10.2217/pgs-2016-0113
Tang H, Wang XS, Huang XP, Roth BL, Butler KV, Kozikowski AP, Jung M, Tropsha A (2009) Novel inhibitors of human histone deacetylase (HDAC) identified by QSAR modeling of known inhibitors, virtual screening, and experimental validation. J Chem Inf Model 49:461–476. https://doi.org/10.1021/ci800366f
Cao GP, Thangapandian S, Son M, Kumar R, Choi YJ, Kim Y, Kwon YJ, Kim HH, Suh JK, Lee KW (2016) QSAR modeling to design selective histone deacetylase 8 (HDAC8) inhibitors. Arch Pharm Res 39(10):1356–1369. https://doi.org/10.1007/s12272-015-0705-5
Pontiki E, Hadjipavlou-Litina D (2012) Histone deacetylase inhibitors (HDACIs). Structure-activity relationships: History and new QSAR perspectives. Med Res Rev 32:1–165. https://doi.org/10.1002/med.20200
Norinder U, Naveja JJ, López-López E, Mucs D, Medina-Franco JL (2019) Conformal prediction of HDAC inhibitors. SAR QSAR Environ Res 30(4):265–277
Nair SB, Teli MK, Pradeep H, Rajanikant GK (2012) Computational identification of novel histone deacetylase inhibitors by docking based QSAR. Comput Biol Med 42(6):697–705. https://doi.org/10.1016/j.compbiomed.2012.04.001
Katritzky AR, Slavov SH, Dobchev DA, Karelson M (2007) Comparison between 2D and 3D-QSAR approaches to correlate inhibitor activity for a series of indole amide hydroxamic acids. QSAR Comb Sci 26:333–345. https://doi.org/10.1002/qsar.200630021
Guo Y, Xiao J, Guo Z, Chu F, Cheng Y, Wu S (2005) Exploration of a binding mode of indole amide analogues as potent histone deacetylase inhibitors and 3D-QSAR analyses. Bioorg Med Chem 13(18):5424–5434. https://doi.org/10.1016/j.bmc.2005.05.016
Xiang Y, Hou Z, Zhang Z (2012) Pharmacophore and QSAR studies to design novel histone deacetylase 2 inhibitors. Chem Biol Drug Des 79:760–770. https://doi.org/10.1111/j.1747-0285.2012.01341.x
Abdel-Atty MM, Farag NA, Kassab SE, Serya RA, Abouzid KA (2014) Design, synthesis, 3D pharmacophore, QSAR, and docking studies of carboxylic acid derivatives as Histone Deacetylase inhibitors and cytotoxic agents. Bioorg Chem 57:65–82. https://doi.org/10.1016/j.bioorg.2014.08.006
Noor Z, Afzal N, Rashid S (2015) Exploration of novel inhibitors for class I histone deacetylase isoforms by QSAR modeling and molecular dynamics simulation assays. PLoS ONE 10(10):e0139588. https://doi.org/10.1371/journal.pone.0139588
Uba AI, Yelekçi K (2017) Identification of potential isoform-selective histone deacetylase inhibitors for cancer therapy: a combined approach of structure-based virtual screening, ADMET prediction and molecular dynamics simulation assay. J Biomol Struct Dyn 21:1–5. https://doi.org/10.1080/07391102.2017.1384402
Dessalew N (2007) QSAR study on amino phenyl benzamides and acrylamides as histone deacetylase inhibitors: an insight into the structural basis of ant proliferative activity. Med Chem Res 16(7–9):449–460. https://doi.org/10.1007/s00044-007-9085-9
Yang JS, Chun TG, Nam KY, Kim HM, Han G (2012) Structure-activity relationship of novel lactam based histone deacetylase inhibitors as potential anticancer drugs. Bull Korean Chem Soc 33:2063–2066. https://doi.org/10.5012/bkcs.2012.33.6.2063
Zhao L, Xiang Y, Song J, Zhang ZA (2013) Novel two-step QSAR modeling work flow to predict selectivity and activity of HDAC inhibitors. Bioorg Med Chem Lett 23(4):929–933. https://doi.org/10.1016/j.bmcl.2012.12.067
Cao GP, Arooj M, Thangapandian S, Park C, Arulalapperumal V, Kim Y, Kwon YJ, Kim HH, Suh JK, Lee KW (2015) A lazy learning-based QSAR classification study for screening potential histone deacetylase 8 (HDAC8) inhibitors. SAR QSAR Environ Res 26:397–420. https://doi.org/10.1080/1062936X.2015.1040453
Liu XH, Song HY, Zhang JX, Han BC, Wei XN, Ma XH, Cui WK, Chen YZ (2010) Identifying novel type ZBGs and nonhydroxamate HDAC inhibitors through a SVM based virtual screening approach. Mol Inf 29:407–420. https://doi.org/10.1002/minf.200900014
Gilson MK, Liu T, Baitaluk M, Nicola G, Hwang L, Chong J (2016) Binding DB in 2015: a public database for medicinal chemistry, computational chemistry and systems pharmacology. Nucleic Acids Res 44:D1045–D1053. https://doi.org/10.1093/nar/gkv1072
Todeschini R, Consonni V, Mauri A, Pavan M (2007) DRAGONs software for the calculation of molecular descriptors, version 5.5 for Windows. Milan, Italy. http://www.talete.mi.it/products/dragon_description.htm
O’Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR (2011) Open Babel: an open chemical toolbox. J Chem Inf 3:1–14. https://doi.org/10.1186/1758-2946-3-33
Mani-Varnosfaderani A, Neiband MS, Benvidi A (2018) Identification of molecular features necessary for selective inhibition of B cell lymphoma proteins using machine learning techniques. Mol divers 12:1–9. https://doi.org/10.1007/s11030-018-9856-x
Farrés M, Platikanov S, Tsakovski S, Tauler R (2015) Comparison of the variable importance in projection (VIP) and of the selectivity ratio (SR) methods for variable selection and interpretation. J Chemom 29(10):528–536. https://doi.org/10.1002/cem.2736
Reis A, Rudnitskaya A, Chariyavilaskul P, Dhaun N, Melville V, Goddard J, Webb DJ, Pitt AR, Spickett CM (2015) Top-down lipidomics of low density lipoprotein reveal altered lipid profiles in advanced chronic kidney disease. J Lipid Res 56:413–422. https://doi.org/10.1194/jlr.M055624
Wang J, Su M, Li T, Gao A, Yang W, Sheng L, Zang Y, Li J, Liu H (2017) Design, synthesis and biological evaluation of thienopyrimidine hydroxamic acid based derivatives as structurally novel histone deacetylase (HDAC) inhibitors. Eur J Med Chem 128:293–309. https://doi.org/10.1016/j.ejmech.2017.01.035
Hu E, Dul E, Sung CM, Chen Z, Kirkpatrick R, Zhang GF, Johanson K, Liu R, Lago A, Hofmann G, Macarron R (2003) Identification of novel isoform-selective inhibitors within class I histone deacetylases. J Pharmacol Exp Ther 307(2):720–728. https://doi.org/10.1124/jpet.103.055541
Melssen W, Wehrens R, Buydens L (2006) Supervised Kohonen networks for classification problems. Chemom Intell Lab Syst 83(2):99–113. https://doi.org/10.1016/j.chemolab.2006.02.003
Vasighi M, Kompany-Zareh M (2013) Classification ability of self organizing maps in comparison with other classification methods. MATCH Commun Math Comput Chem 70:29–44
Omara H, Lazaar M, Tabii Y (2018) Self-organizing maps and principal component analysis to improve classification accuracy. Res J Appl Sci Eng Technol 15(5):190–196. https://doi.org/10.19026/rjaset.15.5851
Ballabio D, Vasighi M (2012) A MATLAB Toolbox for Self Organizing Maps and supervised neural network learning strategies. Chemom Intell Lab 118:24–32
Vapnik VN (1998) Statistical learning theory, 1st edn. Wiley-Interscience, New York. ISBN 978-0-471-03003-4
Chang CC, Lin CJ (2011) LIBSVM: a library for support vector machines. ACM Trans Intell Syst Technol 2:1–27. https://doi.org/10.1145/1961189.1961199
Gramatica P (2007) Principles of QSAR models validation: Internal and external. QSAR Comb Sci 26:694–701. https://doi.org/10.1002/qsar.200610151
Eriksson L, Jaworska J, Worth AP, Cronin MTD, McDowell RM (2003) Methods for reliability and uncertainty assessment and for applicability evaluations of classification- and regression-based QSARs. Environ Health Perspect 111:1361–1375. https://doi.org/10.1289/ehp.5758
Balaban AT (1983) Topological indices based on topological distances in molecular graphs. Pure Appl Chem 5(2):199–206. https://doi.org/10.1351/pac198855020199
Todeschini R, Consonni V, Mannhold R, Kubinyi H, Folkers G (2009) Molecular descriptors for chemoinformatics. Wiley, Weinheim. ISBN 978-3-527-31852-0
Kier LB, Hall LH (1986) Molecular connectivity in structure-activity analysis. Res Stud 33:2096. https://doi.org/10.1002/aic.690331230
Randic M, Kleiner AF, De Alba LM (1994) Distance/distance matrixes. J Chem Inf Comput Sci 34(2):277–286
Bath PA, Poirrette AR, Willett P, Allen FH (1995) The extent of the relationship between the graph–theoretical and the geometrical shape coefficients of chemical compounds. J Chem Inf Comput Sci 35:714–716. https://doi.org/10.1021/ci00026a007
Puzyn T, Leszczynski J, Cronin MT (2010) Recent advances in QSAR studies: methods and applications. Chall Adv Comput Chem Phys 8:1–415. https://doi.org/10.1007/978-1-4020-9783-6
Hall LH, Kier LB, Brown BB (1995) Molecular similarity based on novel atom-type electrotopological state indices. J Chem Inf Comput Sci 35(6):1074–1080
Copeland JC, Zehr LJ, Cerny RL, Powers R (2012) The applicability of molecular descriptors for designing an electrospray ionization mass spectrometry compatible library for drug discovery. Comb Chem high T Scr 15(10):806–815. https://doi.org/10.2174/138620712803901180
Todeschini R, Consoni V (2008) Handbook of molecular descriptors. Methods and principles in medicinal chemistry. Wiley, New York. https://doi.org/10.1002/9783527613106
Fatemi MH, Chahi ZG (2012) QSPR-based estimation of the half-lives for polychlorinated biphenyl congeners. SAR QSAR Environ Res 23(1–2):155–168. https://doi.org/10.1080/1062936X.2011.645876
Suzuki T, Kasuya Y, Itoh Y, Ota Y, Zhan P, Asamitsu K, Nakagawa H, Okamoto T, Miyata N (2013) Identification of highly selective and potent histone deacetylase 3 inhibitors using click chemistry-based combinatorial fragment assembly. PLoS ONE 8(7):68669–68681. https://doi.org/10.1371/journal.pone.0068669
Zhang L, Zhang J, Jiang Q, Zhang L, Song W (2018) Zinc binding groups for histone deacetylase inhibitors. J Enzym Inhib Med Chem 33(1):714–721. https://doi.org/10.1080/14756366.2017.1417274
Jalali-Heravi M, Mani-Varnosfaderani A (2012) Navigating drug-like chemical space of anticancer molecules using genetic algorithms and counter propagation artificial neural networks. Mol Inf 31(1):63–74. https://doi.org/10.1002/minf.201100098
Bertrand P (2010) Inside HDAC with HDAC inhibitors. Eur J Med Chem 45(6):2095–2116. https://doi.org/10.1016/j.ejmech.2010.02.030
Rajak H, Singh A, Raghuwanshi K, Kumar R, Dewangan PK, Veerasamy R, Sharma PC, Dixit A, Mishra P (2014) A structural insight into hydroxamic acid based histone deacetylase inhibitors for the presence of anticancer activity. Curr Med Chem 21(23):2642–2664. https://doi.org/10.2174/09298673113209990191
Bolden JE, Peart MJ, Johnstone RW (2006) Anticancer activities of histone deacetylase inhibitors. Nat Rev Drug Discov 5(9):769–784. https://doi.org/10.1038/nrd2133
Neiband MS, Mani-Varnosfaderani A, Benvidi A (2017) Classification of sphingosine kinase inhibitors using counter propagation artificial neural networks: a systematic route for designing selective SphK inhibitors. SAR QSAR Environ Res 28(2):91–109. https://doi.org/10.1080/1062936X.2017.1280535
Acknowledgements
We gratefully acknowledge the support of this work by Yazd University research council.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Neiband, M.S., Benvidi, A. & Mani-Varnosfaderani, A. Development of classification models for identification of important structural features of isoform-selective histone deacetylase inhibitors (class I). Mol Divers 24, 1077–1094 (2020). https://doi.org/10.1007/s11030-019-10013-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11030-019-10013-0