Abstract
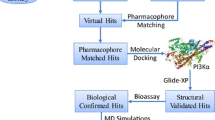
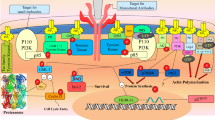
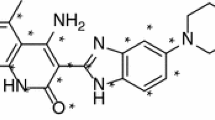
c-KIT is a component of the platelet-derived growth factor receptor family, classified as type-III receptor tyrosine kinase. c-KIT has been reported to be involved in, small cell lung cancer, other malignant human cancers, and inflammatory and autoimmune diseases associated with mast cells. Available c-KIT inhibitors suffer from tribulations of growing resistance or cardiac toxicity. A combined in silico pharmacophore and structure-based virtual screening was performed to identify novel potential c-KIT inhibitors. In the present study, five molecules from the ZINC database were retrieved as new potential c-KIT inhibitors, using Schrödinger’s Maestro 9.0 molecular modeling suite. An atom-featured 3D QSAR model was built using previously reported c-KIT inhibitors containing the indolin-2-one scaffold. The developed 3D QSAR model ADHRR.24 was found to be significant (\(R^{2}=0.9378, Q^{2}=0.7832\)) and instituted to be sufficiently robust with good predictive accuracy, as confirmed through external validation approaches, Y-randomization and GH approach [GH score 0.84 and Enrichment factor (E) 4.964]. The present QSAR model was further validated for the OECD principle 3, in that the applicability domain was calculated using a “standardization approach.” Molecular docking of the QSAR dataset molecules and final ZINC hits were performed on the c-KIT receptor (PDB ID: 3G0E). Docking interactions were in agreement with the developed 3D QSAR model. Model ADHRR.24 was explored for ligand-based virtual screening followed by in silico ADME prediction studies. Five molecules from the ZINC database were obtained as potential c-KIT inhibitors with high in -silico predicted activity and strong key binding interactions with the c-KIT receptor.
Graphical abstract








Similar content being viewed by others
References
Zsebo KM, Williams DA, Geissler EN, Broudy VC, Martin FH, Atkins HL, Hsu RY, Birkett NC, Okino KH, Murdock DC, Jacobsen FW, Langley KE, Smith KA, Takeish T, Cattanach BM, Galli SJ (1990) Stem cell factor is encoded at the SI locus of the mouse and is the ligand for the c-kit tyrosine kinase receptor. Cell 63:213–224. doi:10.1016/0092-8674(90)90302-U
Isakov N, Biesinger B (2000) Lck protein tyrosine kinase is a key regulator of T-cell activation and a target for signal intervention by Herpesvirus saimiri and other viral gene products. Eur J Biochem 267:3413–3421. doi:10.1046/j.1432-1327.2000.01412.x
Drube S, Schmitz F, Gopfert C, Weber F, Kamradt T (2012) C-Kit controls IL-\(1\upbeta \)-induced effector functions in HMC-cells. Eur J Pharmacol 675:57–62. doi:10.1016/j.ejphar.2011.11.035
Robert R Jr (2005) Signaling by Kit protein-tyrosine kinase: the stem cell factor receptor. Biochem Biophys Res Commun 337:1–13. doi:10.1016/j.bbrc.2005.08.055
Kansal N, Silakari O, Ravikumar M (2010) Three dimensional pharmacophore modelling for c-Kit receptor tyrosine kinase inhibitors. Eur J Med Chem 45:393–404. doi:10.1016/j.ejmech.2009.09.013
Wang WL, Healy ME, Sattler M, Verma S, Lin J, Maulik G, Stiles CD, James DG, Johnson BE, Salgia R (2000) Growth inhibition and modulation of kinase pathways of small cell lung cancer cell lines by the novel tyrosine kinase inhibitor STI 571. Oncogene 19:3521–3528. doi:10.1038/sj.onc.1203698
Heinrich MC, Blanke CD, Druker BJ, Corless CL (2002) Inhibition of KIT tyrosine kinase activity: a novel molecular approach to the treatment of KIT-positive malignancies. J Clin Oncol 20:1692–1703. doi:10.1200/JCO.20.6.1692
Imai K, Takaoka A (2006) Comparing antibody and small-molecule therapies for cancer. Nat Rev Cancer 6:714–727. doi:10.1038/nrc1913
Eklund KK (2007) Mast cells in the pathogenesis of rheumatic diseases and as potential targets for anti-rheumatic therapy. Immunol Rev 217:38–52. doi:10.1111/j.1600-065X.2007.00504.x
Jensen BM, Metcalfe DD, Gilfillan AM (2007) Targeting kit activation: a potential therapeutic approach in the treatment of allergic inflammation. Inflamm Allergy Drug Targets 6:57–62. doi:10.2174/187152807780077255
Chen N, Burli RW, Neira S, Hungate R, Zhang D, Yu V, Nguyen Y, Tudor Y, Plant M, Flynn S, Meagher KL, Lee MR, Zhang X, Itano A, Schrag M, Xu Y, Gordon YN, Hu E (2008) Discovery of a potent and selective c-Kit inhibitor for the treatment of inflammatory diseases. Bioorg Med Chem Lett 18:4137–4141. doi:10.1016/j.bmcl.2008.05.089
Gajiwala KS, Wu JC, Christensen J, Deshmukh GD, Diehl W, DiNitto JP, English JM, Greig YH, Jacques SL, Lunney EA, McTigue M, Molina D, Quenzer T, Wells PA, Yu X, Zhang Y, Zou A, Emmett MR, Marshall AG, Zhang HM, Demetri GD (2009) KIT kinase mutants show unique mechanisms of drug resistance to imatinib and sunitinib in gastrointestinal stromal tumor patients. Proc Natl Acad Sci USA 106:1542–1547. doi:10.1073/pnas.0812413106
Force T, Krause DS, Van Etten RA (2007) Molecular mechanisms of cardiotoxicity of tyrosine kinase inhibition. Nat Rev Cancer 7:332–344. doi:10.1038/nrc2106
Zhang L, Zheng Q, Yang Y, Zhou H, Gong X, Zhao S, Fan C (2014) Synthesis and in vivo SAR study of indolin-2-one-based multi-targeted inhibitors as potential anticancer agents. Eur J Med Chem 82:139–151. doi:10.1016/j.ejmech.2014.05.051
Cho TP, Dong SY, Jun F, Hong FJ, Liang YJ, Lu X, Hua PJ, Li LY, Lei Z, Bing H, Ying Z, Qiong LF, Bei FB, Guang LL, Shen GA, Hong SG, Hong SW, Tai MX (2010) Novel potent orally active multitargeted receptor tyrosine kinase inhibitors: synthesis, structure-activity relationships, and antitumor activities of 2-indolinone derivatives. J Med Chem 53:8140–8149. doi:10.1021/jm101036c
Almerico AM, Tutone M, Lauria A (2012) Receptor-guided 3D-QSAR approach for the discovery of c-kit tyrosine kinase inhibitors. J Mol Model 18:2885–2895. doi:10.1007/s00894-011-1304-0
Pan Y, Wang Y, Bryant SH (2013) Pharmacophore and 3D-QSAR characterization of 6-arylquinazolin- 4-amines as Cdc2-like kinase 4 (Clk4) and dual specificity tyrosine-phosphorylation-regulated-kinase 1A (Dyrk1A) inhibitors. J Chem Inf Model 53:938–947. doi:10.1021/ci300625c
Cruciani G, Watson KA (1994) Comparative molecular field analysis using GRID force-field and GOLPE variable selection methods in a study of inhibitors of glycogen phosphorylase b. J Med Chem 37:2589–2601. doi:10.1021/jm00042a012
Ballante F, Ragno R (2012) 3-D QSAutogrid/R: an alternative procedure to build 3-D QSAR models. Methodologies and applications. J Chem Inf Model 52:1674–1685. doi:10.1021/ci300123x
Schuster D, Langer T (2005) The identification of ligand features essential for PXR activation by pharmacophore modeling. J Chem Inf Model 45:431–439. doi:10.1021/ci049722q
Ding L, Tang F, Huang W, Jin Q, Shen H, Wei P (2013) Design, synthesis, and biological evaluation of novel 3-pyrrolo[b]cyclohexylene-2-dihydroindolinone derivatives as potent receptor tyrosine kinase inhibitors. Bioorg Med Chem Lett 23:5630–5633. doi:10.1016/j.bmcl.2013.08.037
Dixon SL, Smondyrev AM, Knoll EH, Rao SN, Shaw DE, Friesner RA (2006) PHASE: a new engine for pharmacophore perception, 3D QSAR model development, and 3D database screening: 1. Methodology and preliminary results. J Comput Aided Mol Des 20:647–671. doi:10.1007/s10822-006-9087-6
Dixon SL, Smondyrev AM, Rao SN (2006) PHASE: a novel approach to pharmacophore modeling and 3D database searching. Chem Biol Drug Des 67:370–372. doi:10.1111/j.1747-0285.2006.00384.x
Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK, Shaw DE, Francis P, Shenkin PS (2004) Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem 47:1739–1749. doi:10.1021/jm0306430
Halgren TA, Murphy RB, Friesner RA, Beard HS, Frye LL, Pollard WT, Banks JL (2004) Glide: a new approach for rapid, accurate docking and scoring. Enrichment factors in database screening. J Med Chem 47:1750–1759. doi:10.1021/jm030644s
Schrödinger Suite 2009 Virtual screening workflow; Glide version 5.5; LigPrep 2.3; QikProp 3.2, Schrödinger, LLC, New York. http://www.schrodinger.com
Roy K, Kar S, Ambure P (2015) On a simple approach for determining applicability domain of QSAR models. Chemom Intell Lab Syst 145:22–29. doi:10.1016/j.chemolab.2015.04.013
Golbraikh A, Shen M, Xiao Z, Xiao Y, Lee K, Tropsha A (2003) Rational selection of training and test sets for the development of validated QSAR models. J Comput Aided Mol Des 17:241–253. doi:10.1023/A:3A1025386326946
Teli MK, Rajanikant GK (2012) Pharmacophore generation and atom based 3D-QSAR of N-iso-propyl pyrrole-based derivatives as HMG-CoA reductase inhibitors. Org Med Chem Lett 2:1–10. doi:10.1186/2191-2858-2-25
Shah UA, Deokar HS, Kadam SS, Kulkarni VM (2010) Pharmacophore generation and atom-based 3D-QSAR of novel 2-(4-methylsulfonylphenyl)pyrimidines as COX-2 inhibitors. Mol Divers 14:559–568. doi:10.1007/s11030-009-9183-3
Golbraikh A, Tropsha A (2002) Beware of \(q2!\). J Mol Graph Mod 20:269–276. doi:10.1016/S1093-3263(01)00123-1
Zhang S, Golbraikh A, Oloff S, Kohn H, Tropsha A (2006) A novel automated lazy learning QSAR (ALL-QSAR) approach: method development, applications, and virtual screening of chemical databases using validated ALL-QSAR models. J Chem Inf Model 46:1984–1995. doi:10.1021/ci060132x
Melagraki G, Afantitis A (2013) Enalos KNIME nodes: exploring corrosion inhibition of steel in acidic medium. Chemom Intell Lab Syst 123:9–14. doi:10.1016/j.chemolab.2013.02.003
Guner O, Henry D (1998) Formula for determining the “goodness of hit lists” in 3D database searches. Accelrys/MDL Information Systems, Inc., San Diego/San Leandro. http://www.netsci.org/Science/Cheminform/feature09.html. Accessed 12 Aug 2015
Chen X, Liu M, Gilson MK (2002) BindingDB: a web-accessible molecular recognition database. Comb Chem High Throughput Screen 4:719–725. doi:10.2174/1386207013330670
Liu T, Lin Y, Wen X, Jorissen RN, Gilson MK (2007) BindingDB: a web-accessible database of experimentally determined protein-ligand binding affinities. Nucleic Acids Res 35(Database issue):198–201. doi:10.1093/nar/gkl999
Chen X, Lin Y, Gilson MK (2001) The binding database: overview and user’s guide. Biopolymers 61:127–141
Compounds: release 4 file series: May 2012. http://cactus.nci.nih.gov/download/nci. Accessed 8 Aug 2015
Melagraki G, Afantitis A (2014) Enalos InSilicoNano platform: an online decision support tool for the design and virtual screening of nanoparticles. RSC Adv 4:50713–50725. doi:10.1039/C4RA07756C
The simple, user-friendly and reliable online application for the AD computation. http://dtclab.webs.com/software-tools or http://teqip.jdvu.ac.in/QSAR_Tools. Accessed 16 Aug 2015
Irwin JJ, Sterling T, Mysinger MM, Bolstad ES, Coleman RG (2012) ZINC: a free tool to discover chemistry for biology. J Chem Inf Model 52:1757–1768. doi:10.1021/ci3001277
Lipinski CA, Lombardo F, Dominy BW, Feeney PJ (2001) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 46:3–26. doi:10.1016/S0169-409X(00)00129-0
Guner OF, Waldman M, Hoffmann RD, Kim JH (2000) Pharmacophore perception, development, and use in drug design, IUL biotechnology series. In: Guner OF (ed) Strategies for database mining and pharmacophore development, 1st edn. International University Line, La Jolla, pp 213–236
Park H, Lee S, Lee S, Hong S (2014) Structure-based de novo design and identification of D816V mutant-selective c-KIT inhibitors. Org Biomol Chem 26:4644–4655. doi:10.1039/c4ob00053f
Wang Y, Xiao J, Suzek TO, Zhang J, Wang J, Zhou Z, Han L, Karapetyan K, Dracheva S, Shoemaker BA, Bolton E, Gindulyte A, Bryant SH (2012) PubChem’s BioAssay database. Nucleic Acids Res 40:400–412. doi:10.1093/nar/gkr1132
Acknowledgments
Authors are thankful to Prof. Sanjay J. Surana, Principal, R. C. Patel Institute of Pharmaceutical Education and Research, Shirpur, India, for availing the facility to carry out the computational work and his valuable support. Authors are also grateful to the North Maharashtra University, Jalgaon, Maharashtra, for providing financial assistance under the scheme “Vice chancellor Research Motivation Scheme (VCRMS),” Sanction No. NMU/11A/VCRMS/Budget-2014-15/Pharmacy-15/170/2015.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of Interest
Corresponding author Prashant J. Chaudhari presented a part of this manuscript at ‘State Level AVISHKAR-2014: The inter-university research convention, Maharashtra’.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Chaudhari, P., Bari, S. In silico exploration of c-KIT inhibitors by pharmaco-informatics methodology: pharmacophore modeling, 3D QSAR, docking studies, and virtual screening. Mol Divers 20, 41–53 (2016). https://doi.org/10.1007/s11030-015-9635-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11030-015-9635-x