Abstract
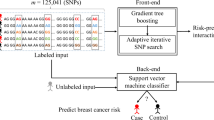
In the view of aggressive nature of Triple-Negative Breast cancer (TNBC) due to the lack of receptors (ER, PR, HER2) and high incidence of drug resistance associated with it, a case–control association study was conducted to identify the contributing genetic risk factors for Triple-negative breast cancer (TNBC). A total of 30 TNBC patients and 50 age and gender-matched controls of Indian origin were screened for 9,00,000 SNP markers using microarray-based SNP genotyping approach. The initial PLINK association analysis (p < 0.01, MAF 0.14–0.44, OR 10–24) identified 28 non-synonymous SNPs and one stop gain mutation in the exonic region as possible determinants of TNBC risk. All the 29 SNPs were annotated using ANNOVAR. The interactions between these markers were evaluated using Multifactor dimensionality reduction (MDR) analysis. The interactions were in the following order: exm408776 > exm1278309 > rs316389 > rs1651654 > rs635538 > exm1292477. Recursive partitioning analysis (RPA) was performed to construct decision tree useful in predicting TNBC risk. As shown in this analysis, rs1651654 and exm585172 SNPs are found to be determinants of TNBC risk. Artificial neural network model was used to generate the Receiver operating characteristic curves (ROC), which showed high sensitivity and specificity (AUC-0.94) of these markers. To conclude, among the 9,00,000 SNPs tested, CCDC42 exm1292477, ANXA3 exm408776, SASH1 exm585172 are found to be the most significant genetic predicting factors for TNBC. The interactions among exm408776, exm1278309, rs316389, rs1651654, rs635538, exm1292477 SNPs inflate the risk for TNBC further. Targeted analysis of these SNPs and genes alone also will have similar clinical utility in predicting TNBC.












Similar content being viewed by others
Abbreviations
- MAF:
-
Minor allele frequency
- OR:
-
Odds ratio
- FISH:
-
Fluorescent in situ hybridization
- nRNA:
-
Non-coding RNA
- GWAS:
-
Genome wide association studies
- DbNSFP:
-
Database for non-synonymous functional prediction
- ER:
-
Estrogen receptor
- PR:
-
Progesterone receptor
- HER2:
-
Human epidermal growth factor receptor 2
- ggplot:
-
Grammar of graphics plot
- PLINK:
-
Population-based linkage analysis
References
Sandhu GS, Erqou S, Patterson H, Mathew A (2016) Prevalence of triple-negative breast cancer in India: systematic review and meta-analysis. J Glob Oncol 2:412–421. doi:10.1200/jgo.2016.005397
Anders CK, Johnson R, Litton J et al (2009) Breast cancer before age 40 years. Semin Oncol 36:237–249. doi:10.1053/j.seminoncol.2009.03.001
Singh M, Jangra B (2013) Association between body mass index and risk of breast cancer among females of north India. South Asian J Cancer 2:121. doi:10.4103/2278-330x.114108
Boyle P (2012) Triple-negative breast cancer: epidemiological considerations and recommendations. Annals of Oncol 23:vi7–vi12. doi:10.1093/annonc/mds187
Mehta S, Shelling A, Muthukaruppan A et al (2010) Predictive and prognostic molecular markers for cancer medicine. Ther Adv Med Oncol 2:125–148. doi:10.1177/1758834009360519
Wang X, Zhao X, Huang X et al (2014) A review of cancer risk prediction models with genetic variants. Cancer Inform. doi:10.4137/cin.s13788
Ross JS, Slodkowska EA (2009) Circulating and disseminated tumor cells in the management of breast cancer. Am J Clin Pathol 132:237–245. doi:10.1309/ajcpji7deolkcs6f
Gunderson KL, Steemers FJ, Lee G et al (2005) A genome-wide scalable SNP genotyping assay using microarray technology. Nat Genet 37:549–554. doi:10.1038/ng1547
Mattos-Arruda LD, Caldas C (2015) Cell-free circulating tumour DNA as a liquid biopsy in breast cancer. Mol Oncol 10:464–474. doi:10.1016/j.molonc.2015.12.001
Karachaliou N, Mayo-de-las-Casas C, Molina-Vila MA, Rosell R (2015) Real-time liquid biopsies become a reality in cancer treatment. Ann Transl Med 3(3):36. doi:10.3978/j.issn.2305-5839.2015.01.16
Steemers FJ, Chang W, Lee G et al (2006) Whole-genome genotyping with the single-base extension assay. Nat Methods 3:31–33. doi:10.1038/nmeth842
Illumina Infinium HD super assay [http://www.illumina.com]
Staaf J, Vallon-Christersson J, Lindgren D et al (2008) Normalization of Illumina Infinium whole-genome SNP data improves copy number estimates and allelic intensity ratios. BMC Bioinform 9:409. doi:10.1186/1471-2105-9-409
Carvalho B, Bengtsson H, Speed TP, Irizarry RA (2006) Exploration, normalization, and genotype calls of high-density oligonucleotide SNP array data. Biostatistics 8:485–499. doi:10.1093/biostatistics/kxl042
Dunning MJ, Barbosa-Morais NL, Lynch AG et al (2008) Statistical issues in the analysis of Illumina data. BMC Bioinformatics 9:85. doi:10.1186/1471-2105-9-85
Zhou T, Li Y, Yang L et al (2016) Silencing of ANXA3 expression by RNA interference inhibits the proliferation and invasion of breast cancer cells. Oncol Rep. doi:10.3892/or.2016.5251
Zeller C, Hinzmann B, Seitz S et al (2003) SASH1: a candidate tumor suppressor gene on chromosome 6q24.3 is downregulated in breast cancer. Oncogene 22:2972–2983. doi:10.1038/sj.onc.1206474
Fu OY, Chang HW, Lin YD et al (2016) Breast cancer-associated high-order SNP-SNP interaction of CXCL12/CXCR4-related genes by an improved multifactor dimensionality reduction (MDR-ER). Oncol Rep. doi:10.3892/or.2016.4956
Dite GS, Macinnis RJ, Bickerstaffe A et al (2015) Breast cancer risk prediction using clinical models and 77 independent risk-associated SNPs for women aged under 50 years: Australian breast cancer family registry. Cancer Epidemiol Biomark Prev 25:359–365. doi:10.1158/1055-9965.epi-15-0838
Liu Y-F, Liu Q-Q, Zhang Y-H, Qiu J-H (2016) Annexin A3 knockdown suppresses lung adenocarcinoma. Anal Cell Pathol 2016:1–9. doi:10.1155/2016/4131403
Ke W, Jiansheng L (2016) Overexpression of ANXA3 is an independent prognostic indicator in gastric cancer and its depletion suppresses cell proliferation and tumor growth. Oncotarget. doi:10.18632/oncotarget.13493
ANXA3 Gene Expressio. http://www.proteinatlas.org/ENSG00000138772-ANXA3/cancer
The R project for statistical computing. http://www.r-project.org
BioConductor. http://www.bioconductor.org
Genome project. http://www.internationalgenome.org/
Zhou T, Li Y, Yang L et al (2017) Annexin A3 as a prognostic biomarker for breast cancer: a retrospective study. Biomed Res Int 2017:1–7. doi:10.1155/2017/2603685
Tong M, Fung T-M, Luk ST et al (2015) ANXA3/JNK signaling promotes self-renewal and tumor growth, and its blockade provides a therapeutic target for hepatocellular carcinoma. Stem Cell Reports 5:45–59. doi:10.1016/j.stemcr.2015.05.013
Pan Q-Z, Pan K, Wang Q-J et al (2015) Annexin A3 as a potential target for immunotherapy of liver cancer stem-like cells. Stem Cells 33:354–366. doi:10.1002/stem.1850
Pénzváltó Z, Tegze B, Szász AM et al (2013) Identifying resistance mechanisms against five tyrosine kinase inhibitors targeting the ERBB/RAS pathway in 45 cancer cell lines. PLoS ONE. doi:10.1371/journal.pone.0059503
Zeller C, Hinzmann B, Seitz S et al (2003) SASH1: a candidate tumor suppressor gene on chromosome 6q24.3 is downregulated in breast cancer. Oncogene 22:2972–2983. doi:10.1038/sj.onc.1206474
Sheyu Lin, Hui Liu, Zhang Junyu Xu, Jiawei Wang Honglian, Qing Sang, Hengwei Zhang, Xuhui Guo, Qinghe Xing, Lin He (2013) Promoter methylation assay of SASH1 gene in breast cancer. JBUON 18(4):891–898
Burgess JT, Bolderson E, Saunus JM et al (2016) SASH1 mediates sensitivity of breast cancer cells to chloropyramine and is associated with prognosis in breast cancer. Oncotarget. doi:10.18632/oncotarget.12020
Ren X, Liu Y, Tao Y et al (2016) Downregulation of SASH1 correlates with tumor progression and poor prognosis in ovarian carcinoma. Oncol Lett. doi:10.3892/ol.2016.4345
O’byrne K, Burgess J, Bolderson E et al (2016) 16P the overexpression of SASH1 stimulates cell death in lung cancer cells. J Thoracic Oncol. doi:10.1016/s1556-0864(16)30130-7
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Aravind Kumar, M., Singh, V., Naushad, S.M. et al. Microarray-based SNP genotyping to identify genetic risk factors of triple-negative breast cancer (TNBC) in South Indian population. Mol Cell Biochem 442, 1–10 (2018). https://doi.org/10.1007/s11010-017-3187-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-017-3187-6