Abstract
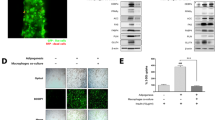
We previously reported that curcumin induces browning of primary white adipocytes via enhanced expression of brown adipocyte-specific genes. In this study, we attempted to identify target proteins responsible for this fat-browning effect by analyzing proteomic changes in cultured white adipocytes in response to curcumin treatment. To elucidate the role of curcumin in fat-browning, we conducted comparative proteomic analysis of primary adipocytes between control and curcumin-treated cells using two-dimensional electrophoresis combined with MALDI-TOF-MS. We also investigated fatty acid metabolic targets, mitochondrial biogenesis, and fat-browning-associated proteins using combined proteomic and network analyses. Proteomic analysis revealed that 58 protein spots from a total of 325 matched spots showed differential expression between control and curcumin-treated adipocytes. Using network analysis, most of the identified proteins were proven to be involved in various metabolic and cellular processes based on the PANTHER classification system. One of the most striking findings is that hormone-sensitive lipase (HSL) was highly correlated with main browning markers based on the STRING database. HSL and two browning markers (UCP1, PGC-1α) were co-immunoprecipitated with these markers, suggesting that HSL possibly plays a role in fat-browning of white adipocytes. Our results suggest that curcumin increased HSL levels and other browning-specific markers, suggesting its possible role in augmentation of lipolysis and suppression of lipogenesis by trans-differentiation from white adipocytes into brown adipocytes (beige).







Similar content being viewed by others
Abbreviations
- 2-DE:
-
Two-dimensional electrophoresis
- ANXA2:
-
Annexin A2
- AMPK:
-
AMP-activated protein kinase
- ATP5B:
-
ATP synthase subunit beta, mitochondrial
- CA3:
-
Carbonic anhydrase 3
- CS:
-
Citrate synthase
- C/EBP/Cebp :
-
CCAAT/enhancer-binding protein/encoding gene
- FABP4:
-
Fatty acid binding protein 4
- HADH:
-
Trifunctional enzyme subunit alpha
- HSL:
-
Hormone-sensitive lipase
- HSP90:
-
Heat shock protein 90 kDa
- PGC-1α:
-
Peroxisome proliferator-activated receptor gamma co-activator 1-alpha
- MDH:
-
Malate dehydrogenase
- PPAR:
-
Peroxisome proliferator-activated receptor
- PRDM16/Prdm16 :
-
PR domain-containing 16/encoding gene
- UCP/Ucp :
-
Uncoupling protein/encoding gene
- WAT:
-
White adipose tissue
References
Haslam DW, James WP (2005) Obesity. Lancet 366:1197–1209
Tonstad S, Despres JP (2011) Treatment of lipid disorders in obesity. Expert Rev Cardiovasc Ther 9:1069–1080
Chang J, Oikawa S, Iwahashi H, Kitagawa E, Takeuchi I, Yuda M, Aoki C, Yamada Y, Ichihara G, Kato M, Ichihara S (2014) Expression of proteins associated with adipocyte lipolysis was significantly changed in the adipose tissues of the obese spontaneously hypertensive/NDmcr-cp rat. Diabetol Metab Syndr 6:8
Noreldin AA, Abd Elhamid AM, Hashem AM, Afifi AM (2013) A pilot study on the use of injection lipolysis in visceral adipose tissues. Aesthet Surg J 33:431–435
Mercer SW, Williamson DH (1988) The influence of starvation and natural refeeding on the rate of triacylglycerol/fatty acid substrate cycling in brown adipose tissue and different white adipose sites of the rat in vivo. The role of insulin and the sympathetic nervous system. Biosci Rep 8:147–153
Ricquier D (1998) Neonatal brown adipose tissue, UCP1 and the novel uncoupling proteins. Biochem Soc Trans 26:120–123
Galmozzi A, Sonne SB, Altshuler-Keylin S, Hasegawa Y, Shinoda K, Luijten IH, Chang JW, Sharp LZ, Cravatt BF, Saez E, Kajimura S (2014) ThermoMouse: an in vivo model to identify modulators of UCP1 expression in brown adipose tissue. Cell Rep 9:1584–1593
Reddy NL, Jones TA, Wayte SC, Adesanya O, Sankar S, Yeo YC, Tripathi G, McTernan PG, Randeva HS, Kumar S, Hutchinson CE, Barber TM (2014) Identification of brown adipose tissue using MR imaging in a human adult with histological and immunohistochemical confirmation. J Clin Endocrinol Metab 99:E117–E121
Harms M, Seale P (2013) Brown and beige fat: development, function and therapeutic potential. Nat Med 19:1252–1263
Cohen P, Spiegelman BM (2015) Brown and beige fat: molecular parts of a thermogenic machine. Diabetes 64:2346–2351
Yoshitomi H, Yamazaki K, Abe S, Tanaka I (1998) Differential regulation of mouse uncoupling proteins among brown adipose tissue, white adipose tissue, and skeletal muscle in chronic beta 3 adrenergic receptor agonist treatment. Biochem Biophys Res Commun 253:85–91
Mulligan JD, Gonzalez AA, Stewart AM, Carey HV, Saupe KW (2007) Upregulation of AMPK during cold exposure occurs via distinct mechanisms in brown and white adipose tissue of the mouse. J Physiol 580:677–684
Bonet ML, Oliver P, Palou A (2013) Pharmacological and nutritional agents promoting browning of white adipose tissue. Biochim Biophys Acta 1831:969–985
Baboota RK, Singh DP, Sarma SM, Kaur J, Sandhir R, Boparai RK, Kondepudi KK, Bishnoi M (2014) Capsaicin induces “brite” phenotype in differentiating 3T3-L1 preadipocytes. Plos One 9:e103093
Zhang Z, Zhang H, Li B, Meng X, Wang J, Zhang Y, Yao S, Ma Q, Jin L, Yang J, Wang W, Ning G (2014) Berberine activates thermogenesis in white and brown adipose tissue. Nat Commun 5:5493
Sellayah D, Bharaj P, Sikder D (2011) Orexin is required for brown adipose tissue development, differentiation, and function. Cell Metab 14:478–490
Shishodia S, Sethi G, Aggarwal BB (2005) Curcumin: getting back to the roots. Ann N Y Acad Sci 1056:206–217
Shehzad A, Ha T, Subhan F, Lee YS (2011) New mechanisms and the anti-inflammatory role of curcumin in obesity and obesity-related metabolic diseases. Eur J Nutr 50:151–161
Lone J, Choi JH, Kim SW, Yun JW (2016) Curcumin induces brown fat-like phenotype in 3T3-L1 and primary white adipocytes. J Nutr Biochem 27:193–202
Joo JI, Oh TS, Kim DH, Choi DK, Wang X, Choi JW, Yun JW (2011) Differential expression of adipose tissue proteins between obesity-susceptible and -resistant rats fed a high-fat diet. Proteomics 11:1429–1448
Forner F, Kumar C, Luber CA, Fromme T, Klingenspor M, Mann M (2009) Proteome differences between brown and white fat mitochondria reveal specialized metabolic functions. Cell Metab 10:324–335
Kamal AH, Kim WK, Cho K, Park A, Min JK, Han BS, Park SG, Lee SC, Bae KH (2013) Investigation of adipocyte proteome during the differentiation of brown preadipocytes. J Proteomics 94:327–336
Birner-Gruenberger R, Susani-Etzerodt H, Waldhuber M, Riesenhuber G, Schmidinger H, Rechberger G, Kollroser M, Strauss JG, Lass A, Zimmermann R, Haemmerle G, Zechner R, Hermetter A (2005) The lipolytic proteome of mouse adipose tissue. Mol Cell Proteomics 4:1710–1717
Okita N, Hayashida Y, Kojima Y, Fukushima M, Yuguchi K, Mikami K, Yamauchi A, Watanabe K, Noguchi M, Nakamura M, Toda T, Higami Y (2012) Differential responses of white adipose tissue and brown adipose tissue to caloric restriction in rats. Mech Ageing Dev 133:255–266
Rodeheffer MS, Birsoy K, Friedman JM (2008) Identification of white adipocyte progenitor cells in vivo. Cell 135:240–249
Kim CY, Le TT, Chen C, Cheng JX, Kim KH (2011) Curcumin inhibits adipocyte differentiation through modulation of mitotic clonal expansion. J Nutr Biochem 22:910–920
Choi DK, Oh TS, Choi JW, Mukherjee R, Wang X, Liu H, Yun JW (2011) Gender difference in proteome of brown adipose tissues between male and female rats exposed to a high fat diet. Cell Physiol Biochem 28:933–948
Kim SW, Park TJ, Choi JH, Aseer KR, Choi JY, Kim YJ, Choi MS, Yun JW (2015) Differential protein expression in white adipose tissue from obesity-prone and obesity-resistant mice in response to high fat diet and anti-obesity herbal medicines. Cell Physiol Biochem 35:1482–1498
Mi H, Muruganujan A, Casagrande JT, Thomas PD (2013) Large-scale gene function analysis with the PANTHER classification system. Nat Protoc 8:1551–1566
da Huang W, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44–57
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25:402–408
Chaudhari HN, Kim SW, Yun JW (2015) Gender-dimorphic regulation of DJ1 and its interactions with metabolic proteins in streptozotocin-induced diabetic rats. J Cell Mol Med 19:996–1009
Wang S, Wang X, Ye Z, Xu C, Zhang M, Ruan B, Wei M, Jiang Y, Zhang Y, Wang L, Lei X, Lu Z (2015) Curcumin promotes browning of white adipose tissue in a norepinephrine-dependent way. Biochem Biophys Res Commun 466:247–253
Liu X, Wu L, Deng G, Li N, Chu X, Guo F, Li D (2008) Characterization of mitochondrial trifunctional protein and its inactivation study for medicine development. Biochim Biophys Acta 1784:1742–1749
Foley JE (1992) Rationale and application of fatty acid oxidation inhibitors in treatment of diabetes mellitus. Diabetes Care 15:773–784
Perez-Perez R, Garcia-Santos E, Ortega-Delgado FJ, Lopez JA, Camafeita E, Ricart W, Fernandez-Real JM, Peral B (2012) Attenuated metabolism is a hallmark of obesity as revealed by comparative proteomic analysis of human omental adipose tissue. J Proteomics 75:783–795
Lindinger PW, Christe M, Eberle AN, Kern B, Peterli R, Peters T, Jayawardene KJ, Fearnley IM, Walker JE (2015) Important mitochondrial proteins in human omental adipose tissue show reduced expression in obesity. J Proteomics 124:79–87
Abe H, Ohtake A, Yamamoto S, Satoh Y, Takayanagi M, Amaya Y, Takiguchi M, Sakuraba H, Suzuki Y, Mori M et al (1993) Cloning and sequence analysis of a full length cDNA encoding human mitochondrial 3-oxoacyl-CoA thiolase. Biochim Biophys Acta 1216:304–306
Kim KB, Lee JW, Lee CS, Kim BW, Choo HJ, Jung SY, Chi SG, Yoon YS, Yoon G, Ko YG (2006) Oxidation-reduction respiratory chains and ATP synthase complex are localized in detergent-resistant lipid rafts. Proteomics 6:2444–2453
Geyik E, Igci YZ, Pala E, Suner A, Borazan E, Bozgeyik I, Bayraktar E, Bayraktar R, Ergun S, Cakmak EA, Gokalp A, Arslan A (2014) Investigation of the association between ATP2B4 and ATP5B genes with colorectal cancer. Gene 540:178–182
McEvily AJ, Mullinax TR, Dulin DR, Harrison JH (1985) Regulation of mitochondrial malate dehydrogenase: kinetic modulation independent of subunit interaction. Arch Biochem Biophys 238:229–236
Zhou SL, Li MZ, Li QH, Guan JQ, Li XW (2012) Differential expression analysis of porcine MDH1, MDH2 and ME1 genes in adipose tissues. Genet Mol Res 11:1254–1259
Carriere A, Jeanson Y, Berger-Muller S, Andre M, Chenouard V, Arnaud E, Barreau C, Walther R, Galinier A, Wdziekonski B, Villageois P, Louche K, Collas P, Moro C, Dani C, Villarroya F, Casteilla L (2014) Browning of white adipose cells by intermediate metabolites: an adaptive mechanism to alleviate redox pressure. Diabetes 63:3253–3265
Harada K, Shen WJ, Patel S, Natu V, Wang J, Osuga J, Ishibashi S, Kraemer FB (2003) Resistance to high-fat diet-induced obesity and altered expression of adipose-specific genes in HSL-deficient mice. Am J Physiol Endocrinol Metab 285:E1182–E1195
Holm C, Osterlund T, Laurell H, Contreras JA (2000) Molecular mechanisms regulating hormone-sensitive lipase and lipolysis. Annu Rev Nutr 20:365–393
Bouwman FG, Wang P, van Baak M, Saris WH, Mariman EC (2014) Increased beta-oxidation with improved glucose uptake capacity in adipose tissue from obese after weight loss and maintenance. Obesity (Silver Spring) 22:819–827
Strom K, Hansson O, Lucas S, Nevsten P, Fernandez C, Klint C, Moverare-Skrtic S, Sundler F, Ohlsson C, Holm C (2008) Attainment of brown adipocyte features in white adipocytes of hormone-sensitive lipase null mice. Plos One 3:e1793
Leggate M, Carter WG, Evans MJ, Vennard RA, Nimmo MA, Sribala-Sundaram S (1985) Determination of inflammatory and prominent proteomic changes in plasma and adipose tissue after high-intensity intermittent training in overweight and obese males. J Appl Physiol 112:1353–1360
Mukherjee R, Kim SW, Park T, Choi MS, Yun JW (2015) Targeted inhibition of galectin 1 by thiodigalactoside dramatically reduces body weight gain in diet-induced obese rats. Int J Obes (Lond) 39:1349–1358
Acknowledgments
This work was supported by the Mid-career Researcher Program (2013R1A2A2A05004195) and SRC Program (Center for Food & Nutritional Genomics, Grant number 2015R1A5A6001906) through a NRF Grant funded by the Ministry of Science, ICT and Future Planning, Korea.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors have declared no conflict of interest.
Rights and permissions
About this article
Cite this article
Kim, S.W., Choi, J.H., Mukherjee, R. et al. Proteomic identification of fat-browning markers in cultured white adipocytes treated with curcumin. Mol Cell Biochem 415, 51–66 (2016). https://doi.org/10.1007/s11010-016-2676-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-016-2676-3