Abstract
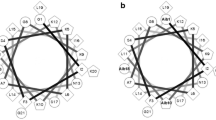
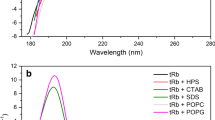
Temporin-SHc (FLSHIAGFLSNLFamide) first isolated from skin extraction of the Tunisian frog Pelophylax saharica, which shows potent antimicrobial activity against Gram-positive bacteria and is highly active against yeasts and fungi without hemolytic activity at antimicrobial concentrations. The peptide adopts well-defined α-helical conformation when bound to SDS micelles. In this study, we explored the effects of residue at position 5 and the N-terminus hydrophobic character on the hydrophilic/polar face of temp-SHc, on its biological activities (antimicrobial and hemolytic) and biophysical properties (hydrophobicity, amphipathicity and helicity). Antibacterial and hemolytic properties of temporin-SHc derivatives depend strongly on physicochemical properties. Therefore, slight decreasing amphipathicity together with hydrophobicity and helicity by the substitution Ile5 → Leu decreased antimicrobial potency approximately twofold without changing of hemolytic activity. It is noteworthy that a conservative amino acid substitution decreases the antimicrobial activity, underlining the differences between Leu/Ile side chains insertion into the lipid bilayer. While the modification of N-terminal hydrophobic character by four residue inversion decreased amphipathicity (twofold) of (4-1)L5temp-SHc and resulted in an increase in antibacterial activity against E. coli, E. faecalis and C. parapsilosis of at least fourfold, its therapeutic potential is limited by its drastic increase of hemolysis (LC50 = 2 μM). We found that the percentage of helicity of temp-SHc analog is directly correlated to its hemolytic activity. Last, the hydrophobic N-terminal character is an important determinant of antimicrobial activity.



Similar content being viewed by others
References
Mangoni ML, Saugar JM, Dellisanti M, Barra D, Simmaco M, Rivas L (2005) Temporins, small antimicrobial peptides with leishmanicidal activity. J Biol Chem 280(984):990. doi:10.1074/jbc.M410795200
Abbassi F, Lequin O, Piesse C, Goasdoué N, Foulon T, Nicolas P, Ladram A (2010) Temporin-SHf a new type of phe-rich and hydrophobic ultrashort antimicrobial peptide. J Biol Chem 285:16880–16992. doi:10.1074/jbc.M109.097204
Grieco P, Luca V, Auriemma L, Carotenuto A, Saviello MR, Campiglia P, Barra D, Novellino E, Mangoni ML (2011) Alanine scanning analysis and structure-function relationships of the frog-skin antimicrobial peptide temporin-1Ta. J Pept Sci 17:358–365. doi:10.1002/psc.1350
Abbassi F, Galanth C, Amiche M, Saito K, Piesse C, Zargarian L, Hani K, Nicolas P, Lequin O, Ladram A (2008) Solution structure and model membrane interactions of temporins-SH antimicrobial peptides from amphibian skin A NMR spectroscopy and differential scanning calorimetry study. Biochemistry 47:10513–11525. doi:10.1021/bi8006884
Mangoni ML, Carotenuto A, Auriemma L, Saviello MR, Campiglia P, Gomez-Monterrey I, Malfi S, Marcellini L, Barra D, Novellino E, Grieco P (2011) Structure-activity relationship conformational and biological studies of temporin L analogues. J Med Chem 54:1298–1307. doi:10.1021/jm1012853
Kamysz W, Mickiewicz B, Rodziewicz-Motowidło S, Greber K, Okrój M (2006) Temporin A and its retro-analogues: synthesis conformational analysis and antimicrobial activities. J Pept Sci 12:533–537. doi:10.1002/psc.762
Yeaman MR, Yount NY (2003) Mechanisms of antimicrobial peptide action and resistance. Pharmacol Rev 55(1):27–55. doi:10.1124/pr.55.1.2
Won A, Pripotnev S, Ruscito A, Ianoul A (2011) Effect of point mutations on the secondary structure and membrane interaction of antimicrobial peptide anoplin. J Phys Chem B 115:2371–2379. doi:10.1021/jp108343g
Abbassi F, Oury B, Blasco T, Sereno D, Bolbach G, Nicolas P, Hani K, Amiche M, Ladram A (2008) Isolation characterization and molecular cloning of new temporins from the skin of the North African ranid Pelophylax saharica. Peptides 29:1526–1533. doi:10.1016/j.peptides.2008.05.008
Abbassi F, Raja Z, Oury B, Gazanion E, Piesse C, Sereno D, Nicolas P, Foulon T, Ladram A (2013) Antibacterial and leishmanicidal activities of temporin-SHd a 17-residue long membrane-damaging peptide. Biochimie 95:388–399. doi:10.1016/j.biochi.2012.10.015
Eisenberg D (1984) Three-dimensional structure of membrane and surface proteins. Annu Rev Biochem 53:595–623. doi:10.1146/annurev.biochem.53.1.595
Chen Y, Mant CT, Farmer SW, Hancock RE, Vasil ML, Hodges RS (2005) Rational design of alpha-helical antimicrobial peptides with enhanced activities and specificity/therapeutic index. J Biol Chem 280:12316–12329. doi:10.1074/jbc.M413406200
Unneberg P, Merelo JJ, Chacón P, Morán F (2001) SOMCD: method for evaluating protein secondary structure from UV circular dichroism spectra. Proteins 42:460–470
Bhunia A, Saravanan R, Mohanram H, Mangoni ML, Bhattacharjya S (2011) NMR structures and interactions of temporin-1Tl and temporin-1Tb with lipopolysaccharide micelles. J Biol Chem 286:24394–24406. doi:10.1074/jbc.M110.189662
Wade D, Silveira A, Silberring J, Kuusela P, Lankinen H (2000) Temporin antibiotic peptides: a review and derivation of a consensus sequence. Protein Pept Lett 7:349–357
Zelezetsky I, Tossi A (2006) Alpha-helical antimicrobial peptides-using a sequence template to guide structure–activity relationship studies. BBA Biomembranes 1758:1436–1449. doi:10.1016/j.bbamem.2006.03.021
Chothia C (1976) The nature of the accessible and buried surfaces in proteins. J Mol Biol 105:1–14
Creighton TE (1983) Proteins structures and molecular properties. Freeman, New York 42
MacCallum JL, Drew-Bennett WF, Tielema DP (2008) Distribution of amino acids in a lipid bilayer from computer simulations. Biophys J 94:3393–3404. doi:10.1529/biophysj.107.112805
Wan CK, Han W, Wu YD (2012) Parameterization of PACE force field for membrane environment and simulation of helical peptides and Helix_Helix association. J Chem Theory Comput 8:300–313. doi:10.1021/ct2004275
Jiang Z, Vasil AI, Hale JD, Hancock RE, Vasil ML, Hodges RS (2008) Effects of net charge and the number of positively charged residues on the biological activity of amphipathic α-helical cationic antimicrobial peptides. Biopolymers 90:369–383. doi:10.1002/bip.20911
Yount NY, Yeaman MR (2005) Immunocontinuum: perspectives in antimicrobial peptide mechanisms of action and resistance. Protein Peptide Lett 12:49–67. doi:10.2174/0929866053405959
Tossi A, Sandri L, Giangaspero A (2000) Amphipathic alpha-helical antimicrobial peptides. Biopolymers 55:4–30. doi:10.2174/0929866053405959
Meijer AB, Spruijt RB, Wolfs CJ, Hemminga MA (2001) Membrane-anchoring interactions of M13 major coat protein. Biochemistry 40: 8815–8820. http://geneura.ugr.es/cgi-bin/somcd/som.cgi?start=1
Conlon JM, Kolodziejek J, Nowotny N (2004) Antimicrobial peptides from ranid frogs: taxonomic and phylogenetic markers and a potential source of new therapeutic agents. BBA Proteins Proteom 1696:1–14. doi:10.1016/j.bbapap.2003.09.004
Acknowledgments
This work was supported by grants from the Centre National de la Recherche Scientifique (CNRS) in France and from the Ministry of Higher Education and Scientific Research in Tunisia.
Conflict of interest
The authors confirm that this article content has no conflicts of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Abbassi, F., Piesse, C., Foulon, T. et al. Effects of residue 5-point mutation and N-terminus hydrophobic residues on temporin-SHc physicochemical and biological properties. Mol Cell Biochem 394, 91–99 (2014). https://doi.org/10.1007/s11010-014-2084-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-014-2084-5