Abstract
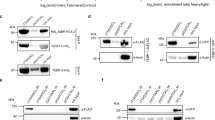
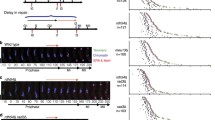
Tankyrases are protein members of the poly(ADP-ribose) polymerase family bearing several ankyrin domain and a WGR domain. They have functional role in telomere maintenance, are found at centrosome, and are associated with vesicular secretion. This diversity in localization and function makes it difficult to identify a unified role for tankyrases. We have shown that the C. elegans orthologue PME-5 is among the most transcriptionally up-regulated genes following ionizing radiations, linking a tankyrase with DNA damage response. Our analysis showed that the up-regulation of PME-5 is translated at the protein level, suggesting an effective role in DNA damage response or DNA repair. In order to gain more information on the potential role of PME-5 in DNA damage response, we analyzed its sub-cellular localization. Using immunostaining as well as gfp reporter assay, we have shown a nuclear localization for PME-5. Moreover, we showed that PME-5 is a ubiquitous nuclear protein expressed throughout the development of the worm and is closely linked to chromatin and condensed chromosomes. Taken together, our data suggest that C. elegans can be used to study the nuclear roles of tankyrase.






Similar content being viewed by others
References
Smith S, de Lange T (2000) Tankyrase promotes telomere elongation in human cells. Curr Biol 10:1299–1302. doi:10.1016/S0960-9822(00)00752-1
Smith S, Giriat I, Schmitt A, de Lange T (1998) Tankyrase, a poly(ADP-ribose) polymerase at human telomeres. Science 282:1484–1487. doi:10.1126/science.282.5393.1484
Lyons RJ, Deane R, Lynch DK, Ye ZS, Sanderson GM, Eyre HJ, Sutherland GR, Daly RJ (2001) Identification of a novel human tankyrase through its interaction with the adaptor protein Grb14. J Biol Chem 276:17172–17180. doi:10.1074/jbc.M009756200
Sbodio JI, Chi NW (2002) Identification of a tankyrase-binding motif shared by IRAP, TAB182, and human TRF1 but not mouse TRF1. NuMA contains this RXXPDG motif and is a novel tankyrase partner. J Biol Chem 277:31887–31892. doi:10.1074/jbc.M203916200
Bae J, Donigian JR, Hsueh AJ (2003) Tankyrase 1 interacts with Mcl-1 proteins and inhibits their regulation of apoptosis. J Biol Chem 278:5195–5204. doi:10.1074/jbc.M201988200
Seimiya H, Smith S (2002) The telomeric poly(ADP-ribose) polymerase, tankyrase 1, contains multiple binding sites for telomeric repeat binding factor 1 (TRF1) and a novel acceptor, 182-kDa tankyrase-binding protein (TAB182). J Biol Chem 277:14116–14126. doi:10.1074/jbc.M112266200
De Rycker M, Price CM (2004) Tankyrase polymerization is controlled by its sterile alpha motif and poly(ADP-ribose) polymerase domains. Mol Cell Biol 24:9802–9812. doi:10.1128/MCB.24.22.9802-9812.2004
Sbodio JI, Lodish HF, Chi NW (2002) Tankyrase–2 oligomerizes with tankyrase-1 and binds to both TRF1 (telomere-repeat-binding factor 1) and IRAP (insulin-responsive aminopeptidase). Biochem J 361:451–459. doi:10.1042/0264-6021:3610451
Kaminker PG, Kim SH, Taylor RD, Zebarjadian Y, Funk WD, Morin GB, Yaswen P, Campisi J (2001) TANK2, a new TRF1-associated poly(ADP-ribose) polymerase, causes rapid induction of cell death upon overexpression. J Biol Chem 276:35891–35899. doi:10.1074/jbc.M105968200
Chang W, Dynek JN, Smith S (2005) NuMA is a major acceptor of poly(ADP-ribosyl)ation by tankyrase 1 in mitosis. Biochem J 391:177–184. doi:10.1042/BJ20050428
Schreiber V, Dantzer F, Ame JC, de Murcia G (2006) Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol 7:517–528. doi:10.1038/nrm1963
Chang P, Coughlin M, Mitchison TJ (2005) Tankyrase-1 polymerization of poly(ADP-ribose) is required for spindle structure and function. Nat Cell Biol 7:1133–1139. doi:10.1038/ncb1322
Chi NW, Lodish HF (2000) Tankyrase is a golgi-associated mitogen-activated protein kinase substrate that interacts with IRAP in GLUT4 vesicles. J Biol Chem 275:38437–38444. doi:10.1074/jbc.M007635200
Yeh TY, Meyer TN, Schwesinger C, Tsun ZY, Lee RM, Chi NW (2006) Tankyrase recruitment to the lateral membrane in polarized epithelial cells: regulation by cell-cell contact and protein poly(ADP-ribosyl)ation. Biochem J 399:415–425. doi:10.1042/BJ20060713
Cook BD, Dynek JN, Chang W, Shostak G, Smith S (2002) Role for the related poly(ADP-Ribose) polymerases tankyrase 1 and 2 at human telomeres. Mol Cell Biol 22:332–342. doi:10.1128/MCB.22.1.332-342.2002
Chiang YJ, Nguyen ML, Gurunathan S, Kaminker P, Tessarollo L, Campisi J, Hodes RJ (2006) Generation and characterization of telomere length maintenance in tankyrase 2-deficient mice. Mol Cell Biol 26:2037–2043. doi:10.1128/MCB.26.6.2037-2043.2006
Hsiao SJ, Poitras MF, Cook BD, Liu Y, Smith S (2006) Tankyrase 2 poly(ADP-ribose) polymerase domain-deleted mice exhibit growth defects but have normal telomere length and capping. Mol Cell Biol 26:2044–2054. doi:10.1128/MCB.26.6.2044-2054.2006
Smith S, de Lange T (1999) Cell cycle dependent localization of the telomeric PARP, tankyrase, to nuclear pore complexes and centrosomes. J Cell Sci 112(Pt 21):3649–3656
Gravel C, Stergiou L, Gagnon SN, Desnoyers S (2004) The C. elegans gene pme-5: molecular cloning and role in the DNA-damage response of a tankyrase orthologue. DNA Repair (Amst) 3:171–182. doi:10.1016/j.dnarep.2003.10.012
Hope I (ed) (1999) C. elegans: a practical approach. Oxford University Press, Oxford
Gagnon SN, Hengartner MO, Desnoyers S (2002) The genes pme-1 and pme-2 encode two poly(ADP-ribose) polymerases in Caenorhabditis elegans. Biochem J 368:263–271. doi:10.1042/BJ20020669
Gautier L, Cope L, Bolstad BM, Irizarry RA (2004) Affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20:307–315. doi:10.1093/bioinformatics/btg405
Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Kristen J, Scherf U, Speed TP (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4:249–264. doi:10.1093/biostatistics/4.2.249
Smyth GK (2004) Linear models and empirical Bayes methods for assessing differential expression in microarray experiments Stat Appl Genet Mol Biol 3: Article 3
Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I (2001) Controlling the false discovery rate in behavior genetics research. Behav Brain Res 125:279–284. doi:10.1016/S0166-4328(01)00297-2
Wettenhall JM, Simpson KM, Satterley K, Smyth GK (2006) affylmGUI: a graphical user interface for linear modeling of single channel microarray data. Bioinformatics 22:897–899. doi:10.1093/bioinformatics/btl025
Luu-The V, Paquet N, Calvo E, Cumps J (2005) Improved real-time RT-PCR method for high-throughput measurements using second derivative calculation and double correction. Biotechniques 38:287–293. doi:10.2144/05382RR05
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. doi:10.1038/227680a0
Harlow E, Lane D (1988) Antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Gartner A, Milstein S, Ahmed S, Hodgkin J, Hengartner MO (2000) A conserved checkpoint pathway mediates DNA damage-induced apoptosis and cell cycle arrest in C. elegans. Mol Cell 5:435–443. doi:10.1016/S1097-2765(00)80438-4
Troemel ER, Chu SW, Reinke V, Lee SS, Ausubel FM, Kim DH (2006) p38 MAPK regulates expression of immune response genes and contributes to longevity in C. elegans. PLoS Genet 2:e183. doi:10.1371/journal.pgen.0020183
Kelly WG, Xu S, Montgomery MK, Fire A (1997) Distinct requirements for somatic and germline expression of a generally expressed Caernorhabditis elegans gene. Genetics 146:227–238
Gisselsson D, Jonson T, Petersen A, Strombeck B, Dal Cin P, Hoglund M, Mitelman F, Mertens F, Mandahl N (2001) Telomere dysfunction triggers extensive DNA fragmentation and evolution of complex chromosome abnormalities in human malignant tumors. Proc Natl Acad Sci USA 98:12683–12688. doi:10.1073/pnas.211357798
Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, Khanna A, Marshall M, Moxon S, Sonnhammer EL, Studholme DJ, Yeats C, Eddy SR (2004) The Pfam protein families database. Nucleic Acids Res 32:D138–D141. doi:10.1093/nar/gkh121
Acknowledgments
We are grateful to Dr. Yuji Kohara for providing us with EST clone yk470c3. We also thank the C. elegans Knock-out consortium for generating the mutant strain pme-5 (ok446), Andrew Fire for the construct pPD118.25 and the Service qRT-PCR du CR-CHUL for quantitative RT-PCR analysis. This work is supported by the Canadian Institutes for Health Research and The Foundation for Research into Children’s Diseases.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
White, C., Gagnon, S.N., St-Laurent, JF. et al. The DNA damage-inducible C. elegans tankyrase is a nuclear protein closely linked to chromosomes. Mol Cell Biochem 324, 73–83 (2009). https://doi.org/10.1007/s11010-008-9986-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-008-9986-z