Abstract
Escherichia coli is one of the simplest hosts which is widely being used to express heterologous proteins. However, without appropriate signal peptide, this host cannot be applied for secretory proteins. Secretory production of recombinant proteins in E. coli has been an issue of interest because of its diverse advantages including cost and time savings, as well as reduction of endotoxin. NS3 from hepatitis C virus (HCV) was chosen as an antigen for vaccine development against HCV virus infections and it connected to gp96 as an adjuvant for stimulating Toll-like receptors (TLRs) to stimulate cytokines secretion by T cells. It was successfully produced in E. coli without using signal peptide previously. In this study, in order to increase the expression level of recombinant NS3-gp96 fusion protein (rNS3-gp96) in periplasmic space, we selected a series of signal peptides. Therefore, to foretell the best signal peptides for expression of NS3-gp96 recombinant protein in E. coli, 52 signal peptides from gram-negative bacteria were chosen and the most important physicochemical features of them were investigated. Therefore, n, h and c regions and signal peptide probability of them were evaluated by signalP software “version 4.1”, and physicochemical features were assessed by ProtParam and PROSO II tools. Eventually, prsK protein, outer membrane pore protein E (phoE), and fimbrial adapter papK protein were determined as the best candidates for the secretory production of rNS3-gp96 in E. coli in our study (with D score 0.899, 0.806, 0.797, respectively). Although, in the experimental investigation, should be considered other influencing parameters.
Similar content being viewed by others
Introduction
Hepatitis C virus (HCV) infection leads to acute and chronic liver diseases in humans such as cirrhosis, chronic hepatitis (Atapour et al. 2017). It is one of the major health problems that has been infected about 200 million people all over the world, and the majority of HCV exposed individuals become steadily unhealthy (Alter et al. 1989). HCV is a single-stranded, RNA Virus that has positive polarity and, is encoded a single open reading frame. Upon translation, the polyprotein is processed by both viral and cellular proteases into individual nonstructural and structural proteins. Although; there is no available vaccine against HCV at the moment HCV, as is one of the protein molecules encoding with RNA, can process into at least ten distinct structural proteins, for instance, C, E1 and E2 and nonstructural proteins such as NS2, NS3, NS4A, NS4B, NS5A and NS5B (Simmonds 2013). Each of them is considered as a potential target for screening of antiviral compounds. Efforts done for developing HCV vaccine have been hindered by several factors including the prone to high-error replication of HCV, lack of suitable animal models and the absence of well-established in vitro knowledge of protective immunity (Singh and Raghava 2001). Novel vaccines are based on molecular technology for eliciting a proper immune response against HCV, including both broadly neutralizing antibodies and effective T-cell response (Naika et al. 2015). Proteins such as NS3, because of stimulation of strong immunity and the existence of conserved epitopes, are attractive for vaccine design; several studies have now shown that T-cell immune responses against NS3 associate with resolution of the infection. Despite the advantages and safety of the recombinant protein vaccines, other strategies to improve their immunogenicity are needed (Pouriayevali et al. 2016). Heat shock proteins (HSPs) facilitate cellular immune responses to antigenic peptides or proteins bound to them. In the present study, we used (HSP gp96) as an adjuvant for creating fusion protein as a candidate vaccine for HCV disease so designed NS3-gp96 fusion protein by connecting the N-terminal NS3 to the N-terminal gp96.
The Prokaryotic system, in particular, Escherichia coli is being employed for production of recombinant protein, in fact, E. coli is one of the best hosts for the expression of recombinant proteins since not only is less expensive but also is very simple to apply (Idicula-Thomas and Balaji 2005; Magnan et al. 2009). Although NS3-gp96 as a recombinant protein can be expressed in E. coli, an important issue is to be considered here; High-level production of functional and soluble recombinant proteins is the major purpose of their expression in bacterial host. Recombinant proteins can be expressed in E. coli as intracellular inclusion bodies; but secretion into the extracellular compartment is a priority as it simplifies downstream purification processes, protects heterologous proteins from proteolysis by cytoplasmic or periplasmic proteases, decrease endotoxin levels and contamination of the product by others host proteins, also improve biological activity and solubility (Gottesman 1996). In E. coli, proteins usually do not secrete into the extracellular compartment except for a few numbers of proteins. Although, small proteins are commonly released into the culture medium depends on the characteristics of signal peptide sequences and proteins (Choi and Lee 2004; Tong et al. 2000). So, we need a tool to direct NS3-gp96 to extracellular compartment of E. coli. In gram-negative bacteria, there are three fate for targeting of expressed protein, including secretion into periplasmic compartment, secretion into outer membrane and extracellular release from outer membrane by common secretory pathway (Desvaux et al. 2004).
The best approach for transfer of rNS3-gp96 to extracellular compartment is using a suitable signal peptide. In fact, in bacteria signal peptides can translocate proteins to periplasmic circumstance by different pathways. In general, there are three main pathways in bacteria for translocation of a secretory protein to periplasmic circumstance that have been classified to the universal secretion pathway (Sec-pathway); the signal recognition particle pathway (SRP pathway) and the twin-arginine translocation (TAT-pathway). Furthermore, among this TAT pathway can transfer folded proteins to periplasmic compartment (Kumari and Chaurasia 2015), whereas Sec and SRP pathways transfer unfolded proteins to periplasmic compartment (De Marco 2009; Natale et al. 2008). Therefore, the researchers are widely using these tools to express secretory protein in which the identification of suitable SP for each protein appears very indispensable to express (De Marco 2009; Gardy and Brinkman 2006; Müller and Bernd Klösgen 2005). There have been some differences particularly in the length and composition of SPs, but in general, any SP is a N-terminal peptide with three key regions; N-terminal region (n-region), a hydrophobic region (h-region) and a cleavable site (c-region). The h-region generally has 7–15 residues while n and c regions have 3–5 residues in length. N and h-regions play a critical role in transferring recombinant proteins into periplasmic space (Emanuelsson et al. 2007; Zimmermann et al. 2011), while c-region plays a vital role as a cleavable site which can be distinguished by signal peptidase enzyme. In spite of SPs key role in the secretion of heterologous proteins, there have been no universal principles to detect them (Emanuelsson et al. 2007; Zhang et al. 2013). In recent decades with the increase in biological tools, biologists are mostly applying method such as machine learning to evaluate the data (Ezziane 2006), as in today, bioinformatics tools have attracted unique attention in biology, because they not only decline the high cost of experiments also provide trustworthy results (Zhang et al. 2013). Our aim was to identify a suitable SP for secretory expression of NS3-gp96 protein in E. coli, therefore, most important features of 52 numbers of SPs from gram-negative bacteria were evaluated and compared using in silico methods and the best of which are introduced for experimental applications.
Materials and Methods
Signal Sequence Collection and Study Design
In this study, amino acid sequences of 52 numbers of SPs were taken from national center of biotechnology information (NCBI) as shown in Table 1. In silico methods such as machine learning techniques were employed to evaluate and characterize the collected signal sequences. Eventually, after trimming and prediction of sub-cellular localization site and also after excluding inappropriate signal peptides, the selected signal peptides were then evaluated to observe whether they have gained high level of secretory expression of rNS3-gp96 protein in E. coli.
In Silico Prediction of n, h and c Regions and Signal Peptide Probability
In order to predict n, h and c regions and signal peptide probability SignalP server version 4.1 (http://www.cbs.dtu.dk/services/SignalP/) was used. These are based on a combination of several artificial neural networks and hidden Markov models (Bendtsen et al. 2004; Petersen et al. 2011). In order to use the server, each SP was connected to N-terminal of NS3-gp96 amino acid sequence and methionine residues were inserted between each SP and NS3-gp96 amino acid sequence.
Physico-Chemical properties and Sub-Cellular Localization of Signal Peptides
In silico study of physicochemical features of signal peptides such as amino acid composition, molecular weight, theoretical PI, Aliphatic Index, solubility index, grand average of hydropathicity (GRAVY) and positively and negatively charged residues were all evaluated by ProtParam server (Walker 2005) (http://web.expasy.org/cgi-bin/protparam/protparam). Prediction of protein solubility upon expression in E. coli was done by the PROSO II software at http://mips.helmholtzmuenchen.de/prosoII. This server uses minute differences between soluble proteins from TargetDB and PDB and undisputedly insoluble proteins from TargetDB, and also literature mining for performing the predictions. In addition, a solubility score between 0 and 1 with a default threshold of 0.6 is given (Smialowski et al. 2012). PROSO II has the maximum prediction accuracy percentage (64.35) compared to some other similar servers, such as CCSOL (54.20), SOLpro (59.95), PROSO (57.85), and recombinant protein solubility (51.4). More importantly, it can be used for heterologous proteins in E. coli (Chang et al. 2013). The solubility tests were performed for SPs linked to rNS3-gp96. In order to sort SPs based on the secretion properties, PRED-TAT server (Bagos et al. 2010) was used (http://www.compgen.org/tools/PRED-TAT/submit). PRED-TAT operates based on hidden Markov models (Bagos et al. 2010). For study of signal peptides sub-cellular location, ProtComp server was used. It merges several methods of protein localization prediction, neural networks-based prediction; direct comparison with updated base of homologous proteins of known localization; and also, comparisons of pentamer distributions calculated for query and DB sequences (http://www.softberry.com). Average accuracy of ProtCompB is 86–100% which depends on space of sub-cellular location, for example, this accuracy in membrane is 100% but in extracellular is 86%. In order to apply PROSO II, PRED-TAT and ProtCompB, each SP was linked to N-terminal of rNS3-gp96 amino acid sequence so that methionine residues were put in between SPs and rNS3-gp96 amino acid sequence (Magnan et al. 2009; Mousavi et al. 2017; Zamani et al. 2015).
Results
In Silico Prediction of n, h and c-Regions and Signal Peptide Probability
The results showed that SPs’ D-scores were between 0.540 (ASPG_ERWCH) and 0.929 (lptA) (Table 2). The most significant parameter for the diagnosis of a SP is the discriminating score (D-score) which is usually described with a cut-off value of 0.5. Actuality only when the SP has a D-score more than 0.50, it is considered. The in silico analysis results of SignalP server has also shown that the highest D-score belonged to lptA, pel2, flgI and ptrA, respectively. Having D-scores < 0.5, Signal peptides AGAR_ALTAT, Lpp and Caf1M were not suitable candidates for the excretion of rNS3-gp96 protein. These signal peptides were deleted among other signal peptides. Then, next analyses were performed on the 49 remaining signal peptides.
As it was mentioned before that n and h regions are important in cleaving SPs from protein, therefore a reliable SP sequence should have obvious n, h and c regions. All of the collected signal peptides have the n-region, h region and c region length between 4 and 11, 8 and 14, and 3 and 13 amino acids respectively. All SP sequences in our study (except three of them) not only had D-score more than 0.50 but also contained obvious n, h and c regions.
Physico-Chemical Properties of Signal Peptides
The in silico results exhibited that the studied SPs length variation was between 17 (dsbG) and 28 (ynfB) amino acid, the lowest and the highest Mw belonged to dsbG (3167.8) and ynfB (2948.7), respectively (Table 3). The results also demonstrated that the range of Net positive charge was between 0 and 4, whereas the range of PI was between 5.75 (ompP) and 12.3 (nrfA). The grand average of hydropathy score (GRAVY) is used to compare SPs overall hydropathy, in fact, this parameter is defined as the sum of hydropathy of amino acids (Zamani et al. 2015). As it is observed the lowest GRAVY belonged to ugpB (0.622) and the highest GRAVY belonged to fecB (2.076). Another factor used to show hydrophobicity is aliphatic index, this factor is defined as the relative volume occupied by aliphatic side chain in an amino acid sequence. According to in silico outcome, the variation in range of aliphatic index was between 79.23 (zraP) and 207.06 (dsbG). Instability index evaluated as another factor too, in general when instability is more than 40, possible proteins is considered unstable, whereas when instability is < 40, it shows the stability of the protein (Zamani et al. 2015). The instability of signal peptides alone and also in connection with rNS3-gp96 was evaluated by instability index. The in silico analysis results showed that the variation in range of instability index was between − 2.6 (papK) and 65.64 (thiB). Instability index of 11 signal peptides including, bla, lamB, appA, appA, ompP, pbpG, phoA, ptrA, thiB, yfeK and Pel2, was more than 40, so they were predicted as unstable. in fact, the analysis results demonstrated that papK (− 2.6) and yhcN (− 2.03) were the most stable signal peptides among the 49 studied signal peptides, respectively (The most unstable signal peptides in connection with rNS3-gp96 were thiB (65.6), appA (60.45) and pbpG (57.99), respectively). The PROSO II server was applied for characterization of rNS3-gp96 solubility in connection with the 49 studied signal peptides. It has been said, solubility of passenger proteins seems essential for secretion, considering that the insoluble proteins tend to aggregate in the inclusion bodies (Baneyx 1999). Considering the solubility of all the tested sequences, this criterion does not look a limiting factor in our analysis, so was not selected as a main decisive factor (Baneyx 1999; Chang et al. 2013). Overexpression of rNS3-gp96 such as other recombinant proteins in E. coli host leads to formation of inclusion body. The inclusion body is a bulk containing the insoluble, nonfunctional and misfolded form of heterologous proteins. To solve this problem, several strategies have been developed. The first is extracellular production of recombinant rNS3-gp96 in E. coli accomplished via attaching signal peptides to N-terminal or C-terminal of gene of interest. The secretory production efficacy of recombinant proteins is different. Therefore, it is essential to assess and evaluate novel signal peptides for optimum selection of proper secretion pathway that is the most effective for the production, processing and secretion of the interested protein (Singh and Panda 2005). The availability of many biological data and advances in computational techniques enable biologist users to study biological systems at different fields from design vaccine to protein engineering, which not only has confidently reduced the time and costs consuming experimental process but has also improved the accuracy of practical studies (Gholami et al. 2015; Zamani et al. 2015). Consequently, the results have indicated that all SPs connected to rNS3-gp96 protein could make a soluble protein, theoretically.
Secretion Sorting and Sub-Cellular Localization
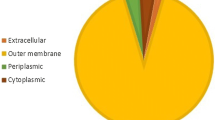
In this study, Sec, SRP and TAT pathways were evaluated by PRED-TAT software and the results revealed that all 49 studied SPs belonged to Sec-pathway. This, in turn, could transfer the expressed rNS3-gp96 recombinant protein to different compartments. Sub-cellular localization analysis showed (by ProtCompB server) that among 49 SPs, 42 SPs can localize rNS3-gp96 in cytoplasm, four SPs can transfer this heterologous protein into extracellular space, and three SPs can localize this heterologous protein into plasma membrane (Table 4).
Discussion
NS3-gp96, as a monomeric protein, lacking disulfide bonds seems a good candidate for secretory production in E. coli. Considering the decisive role of SPs in directing the protein through the membrane, the selection of an appropriate SP is critical. A total number of 52 SPs were selected from several organisms, and their sequences were retrieved from the UniProt server. All 52 numbers of SPs are prokaryotic. Since the native SPs of each host may be more suitable for protein production in that microorganism, 48 SPs were selected from E. coli proteins. Four other SPs from other gram-negative bacteria were also chosen. TAT, Sec and SRP are the main pathways in prokaryote cells directing nascent protein to periplasmic compartment. Furthermore, these pathways operate based on signal peptide recognition, hence it is easily inferred that signal peptides play an important role in folding secretory protein in prokaryote cells (Baneyx and Mujacic 2004; Keller et al. 2012). As mentioned earlier, E. coli is the cheapest and simplest host to express recombinant proteins but the success in using it entirely depends on employing the suitable SPs (Rosano and Ceccarelli 2014). Consequently, the identification of suitable SPs is one of the most vital steps to produce secretory proteins as a recombinant protein in E. coli. Today bioinformatics tools are widely being used in different parts of biological studies largely because they reduce the cost of experiments and they also provide more exact results (Ghasemi et al. 2012; Zamani et al. 2015). As it is observed in this study, it was attempted to employ the most accurate and recent version of bioinformatics tools to predict the variety of SP features. Among various features of SP, net positive charge, aliphatic index, GRAVY, D-score, h-region length, cleavable site and sub-cellular location are more important (Table 5). Accordingly, these features were expected to make the final decision of selecting the best possible SPs. D score is the first parameter in diagnosing an SP, therefore, SPs have all been sorted on the basis of D-score. When D score is more than 0.50, a signal sequence can be considered SP (Zamani et al. 2015). Since all SPs’ D-score in this study is more than 0.50, (except three of them) thereby all of them could be SP but for optimum screening, other features of selection should be considered. N-region is a crucial area in an SP which interferes translocation of a secretory protein, in fact, for maintaining its function, n-region needs a positive charge and this charge is directly linked to the existence of one or more basic residues such as lysine at the beginning of an SP (Zamani et al. 2015). It is believed that switching the basic residues with neutral or acidic residues have an impact on translocation of nascent protein because of the significant role of this positive charge in interacting between SP of nascent protein and membrane phospholipid of RER (Low et al. 2013). As the results show, the variety of net positive charge is considered between 0 and 4, thereby it seems in this stage we do not have enough justification to decide whether to select any SP since all the selected ones have appropriate net positive charge. Another important region which plays a vital role in translocation is h-region, in fact, the most important factor enabling h- region, is hydrophobicity. It has been reported this factor extremely relies on the length of h-region. In fact, the increase in the length of h-region would improve the level of hydrophobicity. Accordingly, there has not been a significant diversity in the length of SPs h-region (9–12) thereby other important factors were used such as aliphatic index and GRAVY in recognition of hydrophobicity. Aliphatic index and GRAVY are the two parameters with direct association with hydrophobicity, in fact, the boost in these parameters, lead to the increase of hydrophobicity (Low et al. 2013; Zamani et al. 2015). As it has been reported in Table 5, among 49 SPs only zraP has low aliphatic index (79.23) and GRAVY (0.746) while in the case of other SPs, no significant difference was observed; therefore, it seems zraP is not a suitable SP to express NS3-gp96 protein. C-region, particularly the three terminal residues that are also named − 3, − 2, − 1 box, are extremely significant in detaching SPs and the secretory proteins after translocation, in fact, − 3, − 2, − 1 boxes are recognized and cleaved by the signal peptidase. Previous studies have indicated that there are typically small or neutral residues such as alanine in − 1 and − 3 positions, whereas there are often big residues in − 2 position which is different with the residues in − 1 and − 3 positions, this residue is illustrated with X (Choi and Lee 2004; Payne et al. 2012; Zamani et al. 2015). As shown in Table 3 all SPs are following this rule and are almost similar to AXA box, therefore we have avoided mentioning this parameter in Table 5. In general the bacteria which uses Sec and SRP pathways translocate unfolded proteins to periplasmic compartment where folding and accumulation are both occurring, on the contrary by the use of TAT pathway they tend to fold secretory proteins in cytoplasm compartment and then translocate the folded proteins to periplasmic compartment for accumulation (De Marco 2009), it seems Sec and SRP pathways are more essential than TAT pathway because folding and purification of secretory proteins in periplasmic or extracellular are easier than in cytoplasm. Since degradation of secretory proteins is less than cytoplasm, it can be concluded that the SPs using these pathways can be more appropriate than SPs which use TAT pathways (Pugsley and Schwartz 1985; Talmadge and Gilbert 1982). As it is shown in Table 4, all SPs in this study belonged to Sec pathway and none could be deleted using this analysis, subsequently other analysis was performed here (it has been reported in previous sections). Finally, it was clarified that among 48 SPs (without zraP), 41 of them can translocate rNS3-gp96 protein to cytoplasmic compartment which could confirm the previous analysis (sec pathway), four SPs could translocate NS3-gp96 to extracellular compartments while three of them translocate rNS3-gp96 protein to membrane compartments. Therefore, it seems only these four signal sequences can be introduced as reliable SP. Therefore, according to D-score (the most important feature), Protein prsK protein, Outer membrane pore protein E (phoE), and Fimbrial adapter papK, were introduced (respectively) as the best signal peptides to express rNS3-gp96 protein into extracellular E. coli. papK which is the most famous signal peptide in this analysis.
Conclusion
Due to existing bioinformatics methods for rapid prediction of functional excretory signal peptides, it is essential to use this approach for effective extracellular production of recombinant proteins in heterologous host. In fact, by selecting an appropriate signal peptide for target protein can be reduce the costs and time of the expression and purification of recombinant proteins. This study evaluated 52 different signal peptides and then selected optimum for secretory production of the recombinant NS3-gp96 protein in E. coli host. This is the first report in theoretical sequence-based analysis of several signal peptides connected with NS3-gp96 and their efficiency in protein secretion to extracellular medium. So, predicting the best SPs by in silico approach would assist biologist and protein engineers to hasten and facilitate the vital projects. Eventually, prsK protein, outer membrane pore protein E (phoE), and fimbrial adapter papK were introduced (respectively) as the best signal peptides to express rNS3-gp96 protein in to extracellular E. coli. Nevertheless, the confirmation of these results needs experimental evaluation.
References
Alter HJ, Purcell RH, Shih JW, Melpolder JC, Houghton M, Choo Q-L, Kuo G (1989) Detection of antibody to hepatitis C virus in prospectively followed transfusion recipients with acute and chronic non-A, non-B hepatitis. New Eng J Med 321:1494–1500
Atapour A, Mokarram P, Mostafavi-Pour Z, Ramezani PA (2017) Molecular cloning, expression, and purification of a recombinant fusion protein (rNT-gp96-NT300). Biopharm Int 30:38–44
Bagos PG, Nikolaou EP, Liakopoulos TD, Tsirigos KD (2010) Combined prediction of Tat and Sec signal peptides with hidden Markov models. Bioinformatics 26:2811–2817
Baneyx F (1999) Recombinant protein expression in Escherichia coli. Curr Opin Biotechnol 10:411–421
Baneyx F, Mujacic M (2004) Recombinant protein folding and misfolding in Escherichia coli. Nat Biotechnol 22:1399
Bendtsen JD, Nielsen H, von Heijne G, Brunak S (2004) Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 340:783–795
Chang CCH, Song J, Tey BT, Ramanan RN (2013) Bioinformatics approaches for improved recombinant protein production in Escherichia coli: protein solubility prediction. Brief Bioinform 15:953–962
Choi J, Lee S (2004) Secretory and extracellular production of recombinant proteins using Escherichia coli. Appl Microbiol Biotechnol 64:625–635
De Marco A (2009) Strategies for successful recombinant expression of disulfide bond-dependent proteins in Escherichia coli. Microb Cell Fact 8:26
Desvaux M, Parham NJ, Scott-Tucker A, Henderson IR (2004) The general secretory pathway: a general misnomer? Trend Microbiol 12:306–309
Emanuelsson O, Brunak S, Von Heijne G, Nielsen H (2007) Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc 2:953
Ezziane Z (2006) Applications of artificial intelligence in bioinformatics: a review. Expert Syst Appl 30:2–10
Gardy JL, Brinkman FS (2006) Methods for predicting bacterial protein subcellular localization. Nat Rev Microbiol 4:741
Ghasemi Y, Dabbagh F, Rasoul-Amini S, Haghighi AB, Morowvat MH (2012) The possible role of HSPs on Behçet’s disease: a bioinformatic approach. Comput Biol Med 42:1079–1085
Gholami A, Shahin S, Mohkam M, Nezafat N, Ghasemi Y (2015) Cloning, characterization and bioinformatics analysis of novel cytosine deaminase from Escherichia coli AGH09. Int J Pept Res Ther 21:365–374
Gottesman S (1996) Proteases and their targets in Escherichia coli. Annu Rev Genet 30:465–506
Idicula-Thomas S, Balaji PV (2005) Understanding the relationship between the primary structure of proteins and its propensity to be soluble on overexpression in Escherichia coli. Protein Sci 14:582–592
Keller R, de Keyzer J, Driessen AJ, Palmer T (2012) Co-operation between different targeting pathways during integration of a membrane protein. J Cell Biol 199:303–315
Kumari S, Chaurasia AK (2015) In silico analysis and experimental validation of lipoprotein and novel Tat signal peptides processing in Anabaena sp. PCC7120. J Microbiol 53:837–846
Low KO, Mahadi NM, Illias RM (2013) Optimisation of signal peptide for recombinant protein secretion in bacterial hosts. Appl Microbiol Biotechnol 97:3811–3826
Magnan CN, Randall A, Baldi P (2009) SOLpro: accurate sequence-based prediction of protein solubility. Bioinformatics 25:2200–2207
Mousavi P, Mostafavi-Pour Z, Morowvat MH, Nezafat N, Zamani M, Berenjian A, Ghasemi Y (2017) In silico analysis of several signal peptides for the excretory production of reteplase in Escherichia coli. Curr Proteom 14:326–335
Müller M, Bernd Klösgen R (2005) The Tat pathway in bacteria and chloroplasts. Mol Memb Biol 22:113–121
Naika HR, Lingaraju K, Chandramohan V, Krishna V (2015) Evaluation of phytoconstituents and molecular docking against NS3 protease of hepatitis C virus. J Pharm Sci Pharmacol 2:96–103
Natale P, Brüser T, Driessen AJ (2008) Sec-and Tat-mediated protein secretion across the bacterial cytoplasmic membrane—distinct translocases and mechanisms. Biochim Biophys Acta Biomemb 1778:1735–1756
Payne SH et al (2012) Unexpected diversity of signal peptides in prokaryotes. MBio 3:e00339–e00312
Petersen TN, Brunak S, von Heijne G, Nielsen H (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Method 8:785
Pouriayevali M-H, Bamdad T, Aghasadeghi M-R, Sadat SM, Sabahi F (2016) Construction and immunogenicity analysis of hepatitis C virus (HCV) truncated non-structural protein 3 (NS3) plasmid vaccine. Jundishapur J Microbiol 9:e33909
Pugsley AP, Schwartz M (1985) Export and secretion of proteins by bacteria. FEMS Microbiol Lett 32:3–38
Rosano GL, Ceccarelli EA (2014) Recombinant protein expression in Escherichia coli: advances and challenges. Front Microbiol 5:172
Simmonds P (2013) The origin of hepatitis C virus. In: Bartenschlager R (ed) Hepatitis C Virus: from molecular virology to antiviral therapy. Springer, Berlin, pp 1–15
Singh SM, Panda AK (2005) Solubilization and refolding of bacterial inclusion body proteins. J Biosci Bioeng 99:303–310
Singh H, Raghava G (2001) ProPred: prediction of HLA-DR binding sites. Bioinformatics 17:1236–1237
Smialowski P, Doose G, Torkler P, Kaufmann S, Frishman D (2012) PROSO II–a new method for protein solubility prediction. FEBS J 279:2192–2200
Talmadge K, Gilbert W (1982) Cellular location affects protein stability in Escherichia coli. Proc Nat Acad Sci USA 79:1830–1833
Tong L et al (2000) Extracellular expression, purification, and characterization of a winter flounder antifreeze polypeptide from Escherichia coli. Protein Expr Purif 18:175–181
Walker JM (2005) The proteomics protocols handbook. Springer, New York
Zamani M, Nezafat N, Negahdaripour M, Dabbagh F, Ghasemi Y (2015) In silico evaluation of different signal peptides for the secretory production of human growth hormone in E. coli. Int J Pept Res Ther 21:261–268
Zhang C, Marcia M, Langer JD, Peng G, Michel H (2013) Role of the N-terminal signal peptide in the membrane insertion of Aquifex aeolicus F1F0 ATP synthase c-subunit. FEBS J 280:3425–3435
Zimmermann R, Eyrisch S, Ahmad M, Helms V (2011) Protein translocation across the ER membrane. Biochim Biophys Acta Biomembr 1808:912–924
Acknowledgements
This study was financially supported by the office of vice-chancellor for research of Shiraz University of Medical Sciences with the Grant No. 95-01-74-11344. The results described in this research were part of PhD student thesis of Shiva Mohammadi.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Mohammadi, S., Mostafavi-Pour, Z., Ghasemi, Y. et al. In silico Analysis of Different Signal Peptides for the Excretory Production of Recombinant NS3-GP96 Fusion Protein in Escherichia coli. Int J Pept Res Ther 25, 1279–1290 (2019). https://doi.org/10.1007/s10989-018-9775-9
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10989-018-9775-9