Abstract
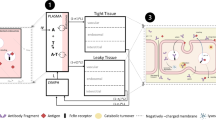
The aim of the present study was to evaluate model identifiability when minimal physiologically-based pharmacokinetic (mPBPK) models are integrated with target mediated drug disposition (TMDD) models in the tissue compartment. Three quasi-steady-state (QSS) approximations of TMDD dynamics were explored: on (a) antibody-target complex, (b) free target, and (c) free antibody concentrations in tissue. The effects of the QSS approximations were assessed via simulations, taking as reference the mPBPK-TMDD model with no simplifications. Approximation (a) did not affect model-derived concentrations, while with the inclusion of approximation (b) or (c), target concentration profiles alone, or both drug and target concentration profiles respectively deviated from the reference model profiles. A local sensitivity analysis was performed, highlighting the potential importance of sampling in the terminal pharmacokinetic phase and of collecting target concentration data. The a priori and a posteriori identifiability of the mPBPK-TMDD models were investigated under different experimental scenarios and designs. The reference model and QSS approximation (a) on antibody-target complex were both found to be a priori identifiable in all scenarios, while under the further inclusion of QSS approximation (b) target concentration data were needed for a priori identifiability to be preserved. The property could not be assessed for the model including all three QSS approximations. A posteriori identifiability issues were detected for all models, although improvement was observed when appropriate sampling and dose range were selected. In conclusion, this work provides a theoretical framework for the assessment of key properties of mathematical models before their experimental application. Attention should be paid when applying integrated mPBPK-TMDD models, as identifiability issues do exist, especially when rich study designs are not feasible.










Similar content being viewed by others
References
Covell DG, Barbet J, Holton OD, Black CD, Parker R, Weinstein JN (1986) Pharmacokinetics of monoclonal immunoglobulin G1, F(ab’)2, and Fab’ in mice. Cancer Res 46:3969–3978
Baxter LT, Zhu H, Mackensen DG, Jain RK (1994) Physiologically based pharmacokinetic model for specific and nonspecific monoclonal antibodies and fragments in normal tissues and human tumor xenografts in nude mice. Cancer Res 54:1517–1528
Garg A, Balthasar JP (2007) Physiologically-based pharmacokinetic (PBPK) model to predict IgG tissue kinetics in wild-type and FcRn-knockout mice. J Pharmacokinet Pharmacodyn 34(5):687–709
Urva SR, Yang VC, Balthasar JP (2010) Physiologically based pharmacokinetic model for T84.66: A monoclonal anti-CEA antibody. J Pharm Sci 99(3):1582–1600
Abuqayyas L, Balthasar JP (2012) Application of PBPK modeling to predict monoclonal antibody disposition in plasma and tissues in mouse models of human colorectal cancer. J Pharmacokinet Pharmacodyn 39:683–710
Shah DK, Betts AM (2012) Towards a platform PBPK model to characterize the plasma and tissue disposition of monoclonal antibodies in preclinical species and human. J Pharmacokinet Pharmacodyn 39:67–86
Nestorov I, Aarons L, Rowland M (1997) Physiologically based pharmacokinetic modeling of a homologous series of barbiturates in the rat: a sensitivity analysis. J Pharmacokinet Biopharm 25(4):413–447
Ito K, Iwatsubo T, Kanamitsu S, Nakajima Y, Sugiyama Y (1998) Quantitative prediction of in vivo drug clearance and drug interactions from in vitro data on metabolism, together with binding and transport. Annu Rev Pharmacol Toxicol 38(1):461–499
Pilari S, Huisinga W (2010) Lumping of physiologically-based pharmacokinetic models and a mechanistic derivation of classical compartmental models. J Pharmacokinet Pharmacodyn 37:365–405
Cao Y, Jusko WJ (2012) Applications of minimal physiologically-based pharmacokinetic models. J Pharmacokinet Pharmacodyn 39:711–723
Cao Y, Balthasar JP, Jusko WJ (2013) Second-generation minimal physiologically-based pharmacokinetic model for monoclonal antibodies. J Pharmacokinet Pharmacodyn 40:597–607
Elmeliegy M, Lowe P, Krzyzanski W (2014) Simplification of complex physiologically based pharmacokinetic models of monoclonal antibodies. AAPS J 16(4):810–842
Fronton L, Pilari S, Huisinga W (2014) Monoclonal antibody disposition: a simplified PBPK model and its implications for the derivation and interpretation of classical compartment models. J Pharmacokinet Pharmacodyn 41:87–107
Levy G (1994) Pharmacologic target-mediated drug disposition. Clin Pharmacol Ther 56(3):248–252
Cao Y, Jusko WJ (2014) Incorporating target-mediated drug disposition in a minimal physiologically-based pharmacokinetic model for monoclonal antibodies. J Pharmacokinet Pharmacodyn 41:375–387
Peletier LA, Gabrielsson J (2012) Dynamics of target-mediated drug disposition: characteristic profiles and parameter identification. J Pharmacokinet Pharmacodyn 39:429–451
Mager DE, Jusko WJ (2001) General pharmacokinetic model for drugs exhibiting target-mediated drug disposition. J Pharmacokinet Pharmacodyn 28(6):507–532
Mager DE, Krzyzanski W (2005) Quasi-equilibrium pharmacokinetic model for drugs exhibiting target-mediated drug disposition. Pharm Res 22(10):1589–1596
Gibiansky L, Gibiansky E, Kakkar T, Ma P (2008) Approximations of the target-mediated drug disposition model and identifiability of model parameters. J Pharmacokinet Pharmacodyn 35:573–591
Ma P (2011) Theoretical considerations of target-mediated drug disposition models: simplifications and approximations. Pharm Res 29:866–882
Eudy R, Riggs MM, Gastonguay MR (2015) A priori identifiability of target-mediated drug disposition models and approximations. AAPS J 17:1280–1284
Grimm HP (2009) Gaining insights into the consequences of target-mediated drug disposition of monoclonal antibodies using quasi-steady-state approximations. J Pharmacokinet Pharmacodyn 36:407–420
Mezzalana E, Lavezzi SM, Zamuner S, De Nicolao G, Ma P, Simeoni M (2015) Integrating target mediated drug disposition (TMDD) into a minimal physiologically based modelling framework: evaluation of different quasi-steady-state approximations. In: PAGE 24, Abstr 3598, Hersonissos, Greece. www.page-meeting.org/?abstract=3598
Padhi D, Jang G, Stouch B, Fang L, Posvar E (2011) Single-dose, placebo-controlled, randomized study of AMG 785, a sclerostin monoclonal antibody. J Bone Miner Res 26(1):19–26
Winkler DG, Sutherland MK, Geoghegan JC, Yu C, Hayes T, Skonier JE, Shpektor D, Jonas M, Kovacevich BR, Staehling-Hampton K, Appleby M, Brunkow ME, Latham JA (2003) Osteocyte control of bone formation via sclerostin, a novel BMP antagonist. EMBO J 22(23):6267–6276
Karlsson J, Anguelova M, Jirstrand M (2012) An efficient method for structural identifiability analysis of large dynamic systems. IFAC Proc Vol 45(16):941–946
Hastie T, Tibshirani R, Friedman J (2008) The elements of statistical learning. Springer, New York, pp 265–267
Hooker A, Staatz CE, Karlsson MO (2007) Conditional weighted residuals (CWRES): a model diagnostic for the FOCE method. Pharm Res 24(12):2187–2197
Bazzoli C, Retout S, Mentré F (2010) Design evaluation and optimisation in multiple response nonlinear mixed effect models: PFIM 3.0. Comput Methods Programs Biomed 98:55–65
Retout S, Comets E, Samson A, Mentré F (2007) Design in nonlinear mixed effects models: optimization using the Fedorov–Wynn algorithm and power of the Wald test for binary covariates. Stat Med 26:5162–5179
Funding
Funding for this analysis was provided by GlaxoSmithKline.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Peiming Ma, Monica Simeoni, and Stefano Zamuner are employed by GlaxoSmithKline and hold company stocks.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Appendix
Appendix
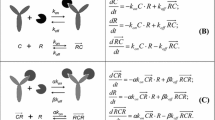
Model equations for the four mPBPK-TMDD models (Full, A, B, and C) are here provided. Parameter and variables notations are as introduced in the sections The full mPBPK-TMDD model and Other mPBPK-TMDD models: quasi-steady-state approximations.
Full Model
Model A
where \(C_{leaky_{free}}\) and \(C\! R_{leaky}\) are computed as:
Model B
where:
Model C
where \(C_{leaky_{free}}\), \(R_{leaky_{free}}\) and \(C\! R_{leaky}\) are obtained as:
Rights and permissions
About this article
Cite this article
Lavezzi, S.M., Mezzalana, E., Zamuner, S. et al. MPBPK-TMDD models for mAbs: alternative models, comparison, and identifiability issues. J Pharmacokinet Pharmacodyn 45, 787–802 (2018). https://doi.org/10.1007/s10928-018-9608-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10928-018-9608-7