Abstract
Our research is motivated by the rapidly-evolving outbreaks of rare and fatal infectious diseases, for example, the severe acute respiratory syndrome (SARS) and the Middle East respiratory syndrome. In many of these outbreaks, main transmission routes were healthcare facility-associated and through person-to-person contact. While a majority of existing work on modelling of the spread of infectious diseases focuses on transmission processes at a community level, we propose a new methodology to model the outbreaks of healthcare-associated infections (HAIs), which must be considered at an individual level. Our work also contributes to a novel aspect of integrating real-time positioning technologies into the tracking and modelling framework for effective HAI outbreak control and prompt responses. Our proposed solution methodology is developed based on three key components – time-varying contact network construction, individual-level transmission tracking and HAI parameter estimation – and aims to identify the hidden health state of each patient and worker within the healthcare facility. We conduct experiments with a four-month human tracking data set collected in a hospital, which bore a big nosocomial outbreak of the 2003 SARS in Hong Kong. The evaluation results demonstrate that our framework outperforms existing epidemic models for characterizing macro-level phenomena such as the number of infected people and epidemic threshold.
Similar content being viewed by others
Introduction
Nosocomial infections, or known as healthcare-associated infections (HAIs), are infections that are acquired in a healthcare setting, such as those caught during an inpatient hospital stay or developed among patients and healthcare staff within a healthcare facility. HAIs have become one of the greatest challenges in the modern world and a global threat to health security. Magill et al. [1] estimated that there were around 722,000 HAIs in U.S. acute care hospitals in 2011, and the [2] reported that around 75,000 patients deaths were related to HAIs. Among the different types of transmission routes of HAIs, the most important and common one is through person-to-person contact, or known as direct-contact transmission. Person-to-person contact transmission takes place when there is a physical contact between an infected or colonized individual and a susceptible person such that disease-causing microorganisms may be transfered. In many cases, due to the incubation period of the infectious disease or unawareness of the disease severity, the disease had already been spread through the hospital before the index patient was identified and quarantined.
A striking example of recent HAI outbreaks through person-to-person contact transmission is the spread of 2013 Middle East respiratory syndrome coronavirus (MERS-CoV) in Saudi Arabia. Assiri et al. [3] reported that, of the 23 confirmed cases of MERS-CoV infection identified in eastern province of Saudi Arabia, 21 cases were acquired by person-to-person transmission in different healthcare facilities including hemodialysis units, intensive care units (ICUs), and in-patient units. In the incident, Patient A with symptoms of dizziness and diaphoresis was admitted to a hospital. No MERS-CoV test was performed and this patient was not suspected of carrying this deadly virus at the time of admission. Another patient, Patient C, was admitted on the next day to the room adjacent to Patient A to undergo hemodialysis. Later, MERS-CoV infection was confirmed in nine other patients who were receiving hemodialysis treatments in the same hospital, six of whom had overlapping time with Patient C undergoing hemodialysis. MERS-CoV infection also developed in a nurse administrator, who had once been present in the ICU where the treatment of Patient A was provided. From this incident, [3] concluded that person-to-person transmission of MERS-CoV can be associated with considerable morbidity in healthcare facilities and suggested that surveillance and infection-control measures are crucial to a global public health response.
Another classic example of large HAI outbreaks is the 2003 severe acute respiratory syndrome (SARS) epidemic in Hong Kong. The 2003 SARS outbreak resulted in 8,096 confirmed cases and 774 deaths in total around the globe [4], and 1755 confirmed cases and 299 deaths in Hong Kong alone (World Health Organization (WHO) 2003). The SARS outbreak began at the Prince of Wales Hospital (PWH) in Hong Kong. The index patient was admitted to PWH before WHO issued a high global alert about cases of this deadly pneumonia. Due to the unawareness of this previously unknown virus, the index patient was not treated as a carrier of a highly infectious and severe disease and SARS started to spread through the hospital among the patients, healthcare workers and visitors, resulting in at least 138 additional cases of SARS acquisition within the hospital (including at least 20 doctors, 34 nurses, 15 allied health workers, and 16 medical students) [5]. The disease was further spread to the community through the individuals leaving the hospital, e.g., leading to the outbreak in a housing estate Amoy Gardens with a total of 329 residents infected and 42 deaths [6], and became an epidemic threatening the world. During the outbreak, the governmental organizations (e.g., Department of Health and Hospital Authority) and public health workers put substantial efforts into contact tracing for effective infection control [6, 7]. It turns out that the contact tracing work was one of the key interventions that helped the Hong Kong Government successfully contained the SARS epidemic, e.g., through screening of symptoms and medical surveillance.
The lesson learned from the 2003 SARS outbreak suggests that a rapid contact tracing is critical for effective quarantine and epidemic management in serious disease outbreaks in hospital settings. A traditional way of conducting contact tracing relies on interviewing. However, conducting interviews may be ineffective because the patient is generally very sick to recall and talk about the past activities and contacts. Another way is to interview the healthcare workers to reconstruct the patients’ past activities. However, both ways cannot guarantee the information is exhaustive and accurate. More importantly, these procedures require lengthy investigations such that prompt isolations cannot be carried out to prevent the disease from spreading.
Our research was motivated by a project initiated at PWH after the SARS outbreak in Hong Kong. The project objectives were to mitigate HAI risk and to support rapid contingency responses in case of severe infectious disease outbreak. Our research team investigated how advanced indoor positioning technologies can be applied to enhance the person-to-person contact traceability for prompt contact tracing, and developed a radio-frequency identification (RFID) system for tracking people’s interactivities (including those among patients and ward staff) and tracing high-risk individuals when infectious disease outbreak occurs. A pilot study was carried out in two medical wards at PWH. Our research team developed an RFID-based real-time locating system for this application. Figure 1 shows a screen-shot of the real-time locating system. For the details of the RFID hardwares, we refer the reader to [8]. It is worth mentioning that the rare experience that the hospital management and staff members of PWH had gone through during the 2003 SARS outbreak and their suggestions were crucial when developing this platform. While our pilot study was enabled by RFID, other indoor positioning technologies are also viable. In our application, RFID was adopted because it is a mature technology and can be deployed at a affordable cost.
A vision-based system is an alternative technology capturing events of person-to-person contacts and monitoring activities of healthcare workers (HCWs) and patients. It can accurately review whether two individuals have been in close contact. Also, most hospitals have installed vision-based systems in the waiting area such as lobbies, escalators, etc., for surveillance purpose. However, such systems may raise privacy issues when installed in wards. In particular, patients’ and HCWs’ privacy is always a primary concern in Hong Kong. It is unlikely that HCWs and patients would accept an environment putting them under constant visual recording. Thus, a sensing method appeared to be more receptive in this study.
To mitigate the risk of HAIs, we characterize the contact patterns among the patients and healthcare workers, and study how an HAI is transmitted through person-to-person contact. From the on-site pilot project conducted at PWH, a large amount of positioning data of patients and healthcare workers were collected. This set of data provides the unique and necessary information to construct dynamic human networks to trace person-to-person contacts, and to develop a more comprehensive understanding of the ways individuals contact with each other in a healthcare environment. After the constructions of human networks within the healthcare facility, we investigate how an HAI is transmitted through person-to-person contact. This investigation requires the capability of tracking the transmission paths of HAIs at an individual level. In other words, we infer the health status of any person at any time and identify potential infected patients and healthcare workers.
Our research is different from traditional epidemiology studies, which investigate the spread of infectious disease among people. Traditional epidemic models, for example, [9,10,11], assume homogeneous human connection (i.e., all individuals in the population are the same) and model the spread of infectious diseases with ordinary differential equations. As suggested by [12], individual differences should be considered when modelling the transmission and designing more effective strategies to reduce the propagation of disease. For HAIs, the assumption that all individuals are equally likely to be infected does not hold, and therefore estimates from community-based models can be inaccurate [13]. Due to the importance of considering individual interactions, some approaches have been proposed to capture the impacts of individual differences using heterogeneous networks, such as the percolation methods [14] and the non-linear dynamical system [15]. However, there is inadequate research on studying the problem of individual-level tracking of HAIs, where most existing methods can hardly be applied to identifying individual infection status. The novelty and uniqueness of our research are as follows. (1) The contact pattern of patients and healthcare workers in a healthcare facility differs from the contact pattern of people in a large region, for example, a city, studied by a majority of the existing epidemic research. A healthcare facility is a relatively closed community with a highly hierarchical and modular structure, leading to dense human connectivity. On the other hand, a city-level population is sparsely connected. (2) We construct time-varying networks to represent dynamic human interaction in a hospital, while many existing models assume static networks that treat human contact unchanged over time. (3) Individual differences in a healthcare setting are related to the roles of individuals in a hospital. As an example, a nurse generally has more interactions with other people than a patient does. (4) We aim to track transmission of HAIs at an individual level that infers the hidden health status for any person at any time, while traditional epidemic models focus on macro-level phenomena such as the total number of infected individuals and epidemic thresholds.
The recent advancement of real-time indoor positioning technologies has provided us with a precious opportunity to study the spread of infectious disease from a new perspective. Given the locational data of patients and healthcare workers in a healthcare institution collected continuously to the system, if an outbreak of an HAI occurs in the institution among patients and healthcare workers, our goal is to track transmission of the HAI at an individual level through person-to-person contact over time-varying contact networks. To achieve the goal, we propose a framework with three key components: time-varying network construction, individual-level transmission tracking and HAI parameter estimation. We realize individual-level transmission tracking with the assumption that all input parameters for the HAI model (HAI parameters) are known. Then we discuss our proposed parameter estimation procedure for effective transmission tracking in a more general setting. Our work leverages a previous RFID tracking study in which we collected locational data of patients and healthcare workers for four months in two medical wards at PWH.
Literature review
Our research investigates the transmission of HAIs over time-varying human networks at an individual level. For an overview of the existing research on epidemic models, we refer the reader to [16,17,18,19,20].
Traditional epidemic models such as the Susceptible-Infected-Recovered (SIR) model and the Susceptible-Infected-Susceptible (SIS) model are based on differential equations [9, 11]. Pairwise models proposed by [21] also belong to this class; in addition, they leverage the local network structure such as the number of pairs or triples to substitute the simple infection number in standard epidemic models. These differential equation-based models have been widely used for analyzing the spread of various types of transmissible diseases. For example, [22] studied the spread of sexually transmitted diseases and considered network heterogeneities in their differential equation-based model. They showed that their predictive scheme was more accurate than models that assume homogeneous networks. Percolation models utilize network degree distributions and epidemic dynamics to examine outcomes of disease outbreaks in the steady state. [14] showed that a large class of standard SIR models could be solved exactly with the use of percolation theory. This approach is more realistic than the traditional approach as it allows heterogeneous and correlated infectiveness times and transmission probabilities. Meyers et al. [23] applied percolation theory to reduce time-consuming calculations and derive different epidemic outcomes on a contact network for the city of Vancouver, British Columbia. Newman [24] and Karrer et al. [25] developed percolation models to study the transmission of two competing diseases based on probability generating functions. Volz [26] showed that SIR model can be represented by a system of ordinary differential equations with the use of probability generating functions. The result enables SIR dynamics to be modeled in random networks.
Another popular class is probabilistic models. This type of models incorporates uncertainty when modeling the disease spreading process. Larson [27] studied the use of “social distancing” (e.g., closing of schools, mandated minimum physical distances between co-workers) to control influenza progression by reducing the frequency and intensity of daily human-to-human contacts. He considered heterogeneous populations, distinguished by high-activity and low-activity persons, in their probabilistic mixing model. Teytelman and Larson [28] considered heterogeneous population where the individuals are different regarding their social activities, pronenesses to infection, and pronenesses to shed virus and spread infection. These attributes determine the rate of human contacts per day and impact the probability that a susceptible individual becomes infected. Yaesoubi and Cohen [29] proposed a discrete-time Markov chain approach to modeling the transmission of diseases. Dynamic optimization techniques can be integrated into their model to aid real-time selection and modification of public health interventions. The studies mentioned above, however, focused on population-level epidemics, which differs from an infectious disease outbreak in a healthcare setting.
As suggested by [30] and [12], human behaviors that might lead to transmission of disease differ significantly between individuals. Much research thus has been carried out to study human interaction pattern. The majority of work in this direction is the use of network analysis. For instance, [31] constructed a bipartite network to represent the connectivity of patients and caregivers in a psychiatric institution. Liljeros et al. [32] studied network properties such as transitivity, assortativity and variation based on a large database constructing from the contact records associated 295,108 inpatients over two years. They concluded that the risk and adverse consequences of epidemic outbreaks might be reduced if these network properties are taken into account when designing the intervention schemes. Ueno and Masuda [33] constructed a hierarchical and modular contact network from a hospital setting. They showed that healthcare workers are main transmitters of diseases and shall be vaccinated with a higher priority. Curtis et al. [34] modeled dynamic contact networks by deriving spatial distributions of healthcare workers and generating random walks to predict human movements in a hospital. Prakash et al. [15] constructed a time-varying network which follows an alternating connectivity behavior to model the day-night pattern of nurse shifts and derived a closed-form equation for the epidemic threshold with their network. In these studies, their main focuses were on the constructions of networks to capture the effects of human interactions on disease transmission. In our work, we construct time-varying dynamic human contact networks from real data and study how these networks can be leveraged for prediction of health status of each individual in a healthcare facility.
Research on disease transmission in a healthcare setting is not adequately addressed in the existing literature. Meyers et al. [31] modelled the disease outbreak in a psychiatric institution, but their model did not utilize real-world human contact data and required some simplified assumptions, e.g., the connection degree of each object in the model follows a Poisson distribution. Prakash et al. [15] used a non-linear dynamical system to model the spread of infections in a time-varying network which takes into account the shifts of nurses in a hospital. Dong et al. [35] modeled nosocomial infections in an MIT college dormitory using graph-coupled hidden Markov models and solved the problem by Gibbs sampling. However, these studies either are limited to describing macro-level epidemic phenomena for HAIs, or did not attempt solutions to the individual-level transmission tracking problem.
Our research studies the transmission of HAIs at an individual level. We propose a modeling framework to describe the interactions between individuals with a dynamic human contact network and tracks infections over this network. Furthermore, we utilize real-time indoor locating technology for tracking the interactions between individuals and constructing their time-varying contact networks. For our modeling contributions, we develop coupled hidden Markov models to solve the problem. Our method captures the difference of the underlying human connectivity where existing epidemic models are unable to address and contributes to more accurate estimations for predicting macro-level outcomes of HAI outbreaks. Moreover, compared to the existing epidemic models, our proposed approach has the unique capability to estimate individual hidden health status and to infer HAI parameters for practical solutions.
Overview
In this section, we introduce the problem of individual-level HAI tracking over dynamic human networks, and then provide an overview of the key components of our proposed solution framework.
Problem definition
Many HAIs are acquired through person-to-person transmission. Our research aims to leverage the use of advanced indoor positioning technologies, such as RFID, for human tracking in healthcare facilities. The availability of the large amounts of locational data about the individuals within the facility, including patients and healthcare workers, enables us to develop a groundbreaking and effective approach to modeling the transmission of disease in a healthcare setting. It provides a new opportunity to characterize person-to-person contacts in a hospital environment and systematically study the transmission pattern of HAIs. More specifically, we aim to develop a modeling framework to track the transmission of HAIs among patients and healthcare workers over time-varying contact networks.
Unlike traditional methods of contact tracing that involve manual processes such as interviewing, advanced positioning technologies collect time-stamped locations of tracked objects automatically and continuously for timely construction of contact networks. In the following, we introduce the terminologies and framework used for our approach.
Definition 1 (Contact)
A pair of individuals is considered to constitute a contact if the distance between them is within a pre-specified distance threshold for a duration that is longer than a pre-specified time threshold.
This definition establishes the foundation that we model the person-to-person transmission of HAIs. A person is considered to have been exposed to the disease if (i) he or she has had any face-to-face contact with an infectious patient, (ii) is in the same hospital room with an infectious patient for more than a certain amount of time, or (iii) is provided care by an infected healthcare worker [3]. With the definition of contact, we construct a person-to-person contact network, denoted by G = (V,E), with vertice set V consisting of individuals in the healthcare facility and edge set E representing the contact records between individuals, i.e., eij ∈ E indicating a contact between individual vi,vj ∈ V. The concept of time-varying contact network can then be extended from the above setting:
Definition 2 (Time-varying contact network)
A time-varying contact network is a series of static contact networks indexed by time points, which is denoted by \(G_{0:T}= \{G_{t}\}_{t = 1}^{T} = \{(V_{t},E_{t})\}_{t = 1}^{T}\), where V t and Et are the sets of individuals and contacts at time t, respectively.
Without loss of generality, we can denote a time-varying contact network by \(G_{0:T} = \{(V,E_{t})\}_{t = 1}^{T}\) because any vertex vk that exists but is not present at time t, that is, vk with \(k \in \{\cup _{s = 1}^{T} V_{s} \}\setminus V_{t}\), can be viewed as an isolated vertex at time t in network Gt. Thus for simplicity, we use \(G_{0:T}=\{(V,E_{t})\}_{t = 1}^{T}\) in the rest of the paper.
In the context of disease, the terms “symptom” and “sign” are both used to refer to an indication of a certain set of medical characteristics that can reflect the presence of a disease, such as runny nose, coughing and fever in a case of influenza. Technically, symptoms and signs are different; a symptom is a feature observed by the patient whereas a sign is observable by the others, e.g., physicians. For consistency, we use observation to represent a symptom or a sign. The presence of a disease, in general, is difficult to be identified without the use of medical diagnostic tests, and therefore, the actual health state of a patient is often hidden. We define the terms observation and health state used for our model as follows.
Definition 3 (Observation)
An observation is a feature that reflects the presence of a disease, which can be a symptom or a sign, or both. \({o}_{t}^{i}\) denotes the observation of person vi at time t. The observation vector \(O_{t}=({o}_{t}^{1},{o}_{t}^{2},\ldots ,{o}_{t}^{n}), n=|V|\) represents the collection of observations of all individuals at time t.
Definition 4 (Health state)
A health state denotes the (hidden) health status of an individual. A patient who is a host of an infectious disease can only be in one health state at a time. \({x}_{t}^{i}\) denotes the hidden health state of person vi at time t. The state vector \(X_{t}=({x}_{t}^{1},{x}_{t}^{2},\ldots ,{x}_{t}^{n}), n=|V|\) denotes the combination of health states of all individuals at time t.
Our objective is to infer the hidden health states of each person at different time points. Observations and networks are necessary to achieve this goal. Observations provide the essential information for diagnoses of an infection, while networks give the information the set of individuals an infected person has contacted. On the other hand, they also impose practical challenges on the accurate identification of hidden health states of a person. Observations can be misleading since the same observation may appear in different health states. The networking effect complicates the problem in the sense that the health state of a person not only is dependent on his/her medical history but also can be affected by other “connected” patients. The challenges motivate us to consider the following overall problem, and the three sub-problems required to tackle in our framework.
Overall problem
Given time-stamped locations of the individuals in a healthcare facility, if an HAI outbreak takes place through person-to-person contact, how can we track the transmission of the HAI over the human contact network at an individual level?
Problem 1 (Time-varying contact network construction)
Given time-stamped locations of the individuals in a healthcare facility, how do we establish contact between any pair of individuals and to construct a time-varying contact network G 0: T to characterize the human connection pattern?
Problem 2 (Individual-level transmission tracking)
Given (1) a time-varying contact network G0:T of the individuals in a healthcare facility, (2) observation vectors O0:T of these people, and (3) the transmission parameter set 𝜃 of the HAI, if an HAI outbreak takes place, how do we identify the hidden health state\({x}_{t}^{i}\) for any person vi at any time t ≥ 0?
Problem 3 (HAI parameter estimation)
Given (1) a time-varying contact network G0:T of the individuals in a healthcare facility, (2) observation vectors O0:T of these people, and (3) a subset 𝜃s of the HAI parameter set 𝜃 with 𝜃s ⊂𝜃, if an HAI outbreak takes place, how do we estimate the unknown parameter set𝜃 ∖𝜃s?
Solution framework
We propose a three-stage solution framework, consisting of the approaches to solving the three respective sub-problems, for tackling the overall problem. An overview of our solution framework is as follows.
- Stage1 :
-
(Time-varying contact network construction) Person-to-person transmission is the primary mode of transmission of HAIs. We define a distance threshold and a time threshold to utilize the time-stamped locations of individuals to construct the list of temporal-spatial co-occurrence events for the establishment of person-to-person contacts. Individuals are divided into multiple groups according to their roles and attributes, and contacts are labeled with four types. We first generate static hierarchical networks based on the types of contacts, and then construct a time-varying contact network.
- Stage2 :
-
(Individual-level transmission tracking) We formulate the problem of individual-level transmission tracking with network-based coupled HMMs, in which the SIS model describes the transmission dynamics of HAIs. HMMs are known to have the power to recover hidden pattern from observable information, and shown to be effective in inference of health states, e.g., [36, 37]. Solutions are obtained in three steps. First, we give basic solutions to a standard HMM by considering all individuals as a single vertex. Then we derive solutions at an individual level by factoring basic solutions according to SIS dynamics. Finally, we reduce the computational complexity of the solutions based on mean-field analysis to speed up computations for large-scale problems.
- Stage3 :
-
(HAI parameter estimation) The problem of HAI parameter estimation is formulated as Maximum Likelihood Estimation (MLE). An auxiliary function is introduced to transform the MLE problem to a computationally efficient optimization problem. By solving this optimization problem with Lagrangian multiplier method, we can obtain the Baum-Welch reestimate and recover the original HAI parameters from this reestimate. This learning method improves the estimation of HAI parameters iteratively.
Table 1 summarizes the notation used throughout this paper.
Time-varying contact network construction
Contact establishment
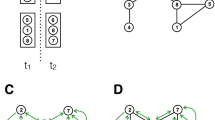
We can establish a linkage between two persons in the contact network by utilizing the information about their temporal-spatial co-occurrence. First, movement trajectories of individuals, e.g. trajectories of person vi and person vj shown in Fig. 2a, are extracted from the time-stamped locational data captured from the indoor positioning infrastructure. The distance between person vi and person vj at any time is calculated; examples of the distances are represented by dashed lines in Fig. 2a. Let dij(t) be a function of time t that measures the distance between persons vi and vj. A contact between vi and vj is established if ∃t1,t2, where t2 − t1 > ΔTth such that
where Dth is the distance threshold and ΔTth is the time threshold.
Static hierarchical network generation
A three-level hierarchical network is generated to represent the human connectivity in a hospital environment. Individuals are labeled with their role classes and divided into a patient group and a caregiver group. The patient group is further divided according to the ward where a patient is staying. Correspondingly, there are four types of contacts, namely, intra-ward contacts, inter-ward contacts, intra-type contacts and inter-type contacts. An example of a simple hierarchical contact network is shown in Fig. 3. The bottom-level network consists of patients and they are linked according to intra-ward contacts; the middle-level network consists of both patients and caregivers and they are linked with intra-type and inter-ward contacts; the top-level network consists of inter-type contacts between patient-caregiver pairs.
Time-varying contact network construction
A time-varying network can be represented as a series of static networks. We divide a continuous period into discrete time intervals and construct the time-varying contact network by combining the static networks constructed for each time period in sequence. In a hospital setting, it is common to divide a day into a daytime nighttime sessions [15], because most healthcare workers have fixed shift times and human connectivity has dissimilar structures in the two sessions.
Most healthcare facilities adopt a hierarchical and modular structure for the separation of wards and different roles of individuals. Interaction pattern differs between the intra-ward and inter-ward contacts or between the intra-type and inter-type contacts. The time-varying hierarchical contact network naturally captures such characteristics in a hospital environment. Moreover, the hierarchical network is an extension of the bipartite patient-caregiver network in [31]. The bipartite patient-caregiver network assumes that only inter-type contacts exist, but does not consider contacts within the same individual group. Undoubtedly, it would be more practical to consider contacts among caregivers as they are the main transmitters of HAIs.
Individual-level transmission tracking
Transmission dynamics
The classical SIS model formulates transmission dynamics of HAIs. There are two health states in the SIS model, namely, susceptible and infected. A person can be either susceptible or infected at one time. A susceptible person vi might get infected with an infection rate τi due to a contact with an infectious patient. An infected patient vj gets recovered independently with a recovery rate μj, and turns from the infected state to the susceptible state immediately. Let health state \({x}_{t}^{i}\) be a binary variable such that \({x}_{t}^{i}= 1\) and 0 respectively indicate that person vi is infected and suspectible at time t. Let \(\mathcal {N}_{t}(v_{i})\) be the neighbor set of vi at time t, where a neighbor of vi is defined to be the set of adjacent vertices of vi on graph Gt. In the SIS model, the transmission through person-to-person contact is determined by the following set of equations.
Equation 1 represents the case that a susceptible person vi remains susceptible at the next time period if all his/her infectious neighbors fail to transmit the infection to vi. Equation 2 indicates that a susceptible person gets infected if one or more of the infectious neighbors transmit the infection to him/her successfully. Equations 3 and 4 respectively state that an infected person recovers and remains infected independently. The SIS dynamics is shown in Fig. 4. The links between vertices represent the person-to-person contacts. At time t, v1,v2 and v3 are infected while v4 and v5 are susceptible. At time t + 1, v4 remains susceptible as the infected neighbors v1 and v3 both fail to transmit the disease to v4. v5 gets infected because v1 transmits the infection to v5 successfully. v2 recovers independently and becomes susceptible at time t + 1 , while v1 and v3 remain infected.
Detection, tracing and prediction
The problem of individual-level transmission tracking is to identify the hidden health state of a person at any time, given the set of observations, parameters of the HAI model, and the contact networks. Suppose that the current time is t. Specifically, the problems of tracing, detection and prediction are respectively to infer the hidden state of an individual before, at and after time t, as illustrated in Fig. 5. Formally, given the observation set O0:t, the time-varying contact network G0:t, and the parameter set 𝜃 of an HAI, the problem of individual-level transmission tracking is to determine a mapping function
We call the function f for detection if s = t, tracing if s < t, and prediction if s > t.
We propose network-based coupled HMMs to tackle the individual-level transmission tracking problem. As shown in Fig. 6, a standard HMM has two sequences of components: a sequence of hidden states of a Markov chain and a sequence of observations. An observation is dependent on its hidden state only, but not affected by other states and observations. HMMs have the power to reveal hidden pattern from observable information and have been widely applied in various fields such as speech recognition and social network analysis. The coupled HMMs incorporate several sub-HMMs together under a network structure, as illustrated in Fig. 7. This connection creates the interdependence of the hidden states of multiple sub-HMMs. For example, the hidden state of HMM-2 at time t is not only determined by its own state at time t − 1, but also affected by the hidden states of HMM-1 and HMM-3 at time t − 1.
We propose a two-phase approach to deriving solutions from the coupled HMMs. First, the coupled HMMs are regarded as a standard HMM to give basic solutions to the problems of detection, tracing and prediction. In the standard HMM, all individuals are considered as single vertices such that the combined state Xt is determined rather than individual \({x}_{t}^{i}\). The second phase is to derive solutions for individual-level transmission tracking based on factorization and mean-field analysis.
For each person vi, we denote the detection probability at time t by \({\pi }_{t}^{i}\), the tracing probability at time s by \({\pi }_{s|t}^{i}\), and the k-step ahead prediction probability at time t + k by \({\pi }_{t+k|t}^{i}\). Correspondingly, πt, πs|t and πt + k|t respectively denote the detection probability at time t, the tracing probability at time k and the k-step ahead prediction probability at time t + k. Let ht|t− 1 = P(Xt|Xt− 1) be the state transition probability and ϕt = P(Ot|Xt) be the observation probability. A standard HMM provides the detection, tracing and prediction probabilities in recursive forms by the following set of equations [38], respectively:
where \(\beta _{s|t} = {\sum }_{X_{s + 1}} \beta _{s + 1|t} \phi _{s + 1} h_{s + 1|s}\) can be computed with a backward approach. The basic solutions can be obtained by the analogies of detection, tracing and prediction to filtering, smoothing and prediction in a standard HMM.
However, treating all individuals as a single vertex is incapable of tracking the health state of each individual. It is also impractical to apply the basic solutions directly due to the high computational complexity. For example, if we are to track the transmission of an SIS-type HAI among n individuals, the computational complexities for the detection and tracing procedures are respectively O(22n) and O(2n). Thus, we propose an integrated approach of factorization and mean-field analysis to reducing the complexities.
Detection
Since the basic solutions obtained from a standard HMM treat all individuals as a whole, we first factorize the basic solutions for individual-level detection, and then reduce the computational complexity using mean-field analysis. In this way, we can solve the individual-level detection problem and substantially improve the solvability for large-scale problems.
Assumption 1 (Independence assumption)
The one-step-ahead prediction probability of a sub-HMM is independent of those of other sub-HMMs, or formally
where n is the number of sub-HMMs.
The state transition probability ht|t− 1 is determined by SIS dynamics. By Eqs. 5, 6, and 7, the individual states are conditionally independent, that is,
Theorem 1
The solution to the problem of individual-level detection is given by
and the basic solution to the detection problem can be factored in a product form
Factorization of the basic solution gives the detection probability for each individual. The complexity is reduced to O(2n) after factorization, and the computational burden now becomes the calculation of individual-level one-step-ahead prediction probability \(\pi _{t|t-1}^{i}\). We apply mean-field analysis for solving \( \pi _{t|t-1}^{i} \) to reduce the overall computational complexity further. The mean-field analysis studies the behavior of a large and complex system in view of simpler systems. Such system considers a large number of small individuals who interact with each other, where the effects of the other individuals on any given individual can be approximated by an averaged effect. With independence assumptions and decomposition, mean-field analysis reduces a multiple-body problem to a one-body problem.
Theorem 2
The solution to the problem of individual-level one-step-ahead prediction is given by
where \( p_{t|t-1}^{i} = \prod \nolimits _{j:v_{j} \in \mathcal {N}_{t-1} (v_{i})}\! \left (\pi _{t-1}^{j} (1) \!\cdot (1 - \tau _{i}) + \pi _{t-1}^{j}(0) \right ) \) .
Theorems 1 and 2 enable us to recursively calculate the one-step-ahead prediction and detection probabilities for each individual. The computational complexity is, therefore, reduced to O(n2) by mean-field analysis.
Algorithm 1 outlines the forward algorithm for solving individual-level detection. Let detection vector \( \boldsymbol {\pi }_{t}^{i} = \left ({\pi _{t}^{i}}(0), {\pi _{t}^{i}}(1)\right ) \), observation vector \( \boldsymbol {\phi }_{t}^{i} = \left ({\phi }_{t}^{i}({O_{t}^{i}}|0), {\phi }_{t}^{i}({O_{t}^{i}}|1)\right ) \), and ρi be the initial distribution of hidden states of individual vi. We initialize and calculate the detection probability for each individual at time 0. Then we recursively compute the one-step-ahead probability and the detection probability.

Tracing
The basic solution to the tracing problem in the standard HMM requires a backward computing procedure for βs|t,s < t. The procedure introduced in Sub-section appears to be not applicable for tracing because βs|t is not normalized. To derive the tracing probability at an individual level, we rewrite the basic solution based on the following formula [39, 40]:
Theorem 3
The solution to the problem of individual-level tracing is provided by
and the basic solution to the tracing problem can be factored in the following product form:
where s ≤ t, \( P(x_{s + 1}^{i}= 0|{\pi _{s}^{i}}= 1)=\mu _{i} \), and \( P(x_{s + 1}^{i}= 1|{\pi _{s}^{i}}= 1)= 1-\mu _{i} \).
Algorithm 2 outlines the forward-backward algorithm for solving individual-level tracing. We first implement the forward algorithm to obtain the detection probability and the one-step-ahead probability. Then we set the tracing probability equal to the detection probability at time T. Thus, the tracing probability with time earlier than T can be computed recursively in a backward fashion.

Prediction
We have provided the solution approach to the problem of one-step-ahead prediction in the previous sub-section. Trivially, we can solve the problem of s-step ahead prediction by substituting the one-step transition probability with the s-step transition probability. Here we introduce the pure prediction probability \( \pi _{t|0}^{i}(x) \). In pure prediction, no health observations are given, but the initial outbreak information is available. The health states of individuals are completely determined by the initial conditions and epidemic dynamics given by the SIS model. Intuitively, the effect of having no observation at all is equivalent to the setting that all individuals have the same observation at any time. Based on this intuition, we modify the computation for the detection probability \( {\pi _{t}^{i}}(x) \) to derive \( \pi _{t|0}^{i}(x) \). We set the observation space to {0} and the observation probability ϕ(⋅,0) to one. By doing so, this tracking procedure becomes pure prediction. We calculate the tracking probability \( {\pi _{t}^{i}}(x) \) under this condition, and we have \( \pi _{t|0}^{i}(x)={\pi _{t}^{i}}(x) \) with ϕ(⋅,0) = 1. Pure prediction is consistent with the non-linear dynamical system (NLDS) discussed in [15]. The model reduces to NLDS when observable features of HAIs are unavailable. As observations provide useful information for more effective estimation of the health states, one-step-ahead prediction is expected to give better performance than the NLDS or pure prediction.
HAI parameter estimation
In “Individual-level transmission tracking”, we discussed our modeling framework on tracking HAI transmission under the assumption that all the required parameters – the infection rate, the recovery rate, the initial state distribution and the observation probability matrix – are given. In practice, however, the real values of these parameters are very likely not known exactly. In general, at the beginning of an outbreak, making an initial guess is the only possible option as no prior information is available. In this section we present an estimation method to refine the guess in a step-by-step manner, thus guaranteeing the practicality of our approach for real-world problems. Let \( \boldsymbol {\theta } = (\{ \rho ({x_{0}^{i}}) \}, \{ \tau _{i} \}, \{ \mu _{i} \}, \{ {\phi _{x}^{o}} \}) \) be an HAI parameter configuration. The goal of HAI parameter estimation is to find the best 𝜃 that maximizes the likelihood function \(\mathcal {L}(\boldsymbol {\theta }) = P(O_{0:T}|\boldsymbol {\theta }) \),
As analytical global optimal solutions are unlikely to exist, we use the Baum-Welch method [41, 42] to solve the problem. Let λ = ({ρ(X)},{h(X,X′)},{ϕ(X,O)}). We first consider the coupled HMMs as a single HMM for estimating λ, and then recover the original parameter set 𝜃 from λ. We introduce the auxiliary function
Using Jensen’s inequality, we obtain
Let \( \boldsymbol {\lambda }^{*}=\arg \max \limits _{\bar {\boldsymbol {\lambda }}} Q(\boldsymbol {\lambda }, \bar {\boldsymbol {\lambda }}) \). we have
The above fact suggests that the likelihood of λ never exceeds the likelihood of λ∗ and that computing a new estimate \( \bar {\boldsymbol {\lambda }} \) by maximizing the auxiliary function \( Q(\boldsymbol {\lambda },\bar {\boldsymbol {\lambda }}) \) improves the likelihood. Thus, we can derive a maximum likelihood estimate by iteratively updating \( \bar {\boldsymbol {\lambda }} \) until convergence. In this way, the original intractable optimization problem is reduced to maximizing the auxiliary function \(Q(\boldsymbol {\lambda },\bar {\boldsymbol {\lambda }}) \), which can be solved with a Lagrangian multiplier approach efficiently.
Consider the following optimization problem
where \( \mathcal {X} \) is the value set of state vector X.
By substituting
we can write the Lagrangian function as
where η,γj and ωj are Lagrangian multipliers. The optimization problem thus becomes
which can be separated to three independent maximization problems. Solving each sub-problem, we have
where πt,t+ 1|T(X,X′) = P(Xt = X,Xt+ 1 = X′|O0:T) denotes the probability that the node is at state X at time t and at state X′ at time t + 1. As an example, we take the derivative of \( \bar {h}(X,X^{\prime }) \) to obtain the solution to the maximization problem. By letting \( L_{3} \! = \! \sum \limits _{X_{0:T}} P(X_{0:T}|O_{0:T}, \boldsymbol {\lambda }) \sum \limits _{t = 0}^{T-1} \log \bar {h}({X}_{t}, {X}_{t + 1}) + \sum \limits _{j = 1}^{|\mathcal {X}|} \omega _{j} \left (\sum \limits _{X^{\prime }\in \mathcal {X}} \bar {h}(X,X^{\prime }) - 1 \right ) \) and \( X_{0:T}^{\prime }=(X_{0:T}: X_{t}=X,X_{t + 1}=X^{\prime }) \), we have
By setting \(\frac {\partial L_{3}}{\partial \bar {h}(X,X^{\prime })} = 0\), we have
By setting \( \omega _{j} = -{\sum }_{t = 0}^{T-1} \pi _{t|T}(X) \), the third constraint is satisfied, and Equality (20) holds.
Note that \( \bar {\rho }(X_{0}), \bar {\phi }(X,F) \text { and } \bar {h}(X,X^{\prime }) \) in Eqs. 18, 19 and 20 are the same with the Baum-Welch reestimate. Iteratively updating \( \bar {\boldsymbol {\lambda }} \) in this manner keeps improving the estimate of λ until it converges. The subsequent step to recover the original parameter configuration \( \bar {\boldsymbol {\theta }} \) from \( \bar {\boldsymbol {\lambda }} \). As an example, we illustrate the recovery of SIS parameters τi and μi.
Lemma 1
The probability πt,t+ 1|T(X,X′) can be derived from the detection probability πt(X), the tracing probability πt+ 1|T(X′), the one-step-ahead prediction probability πt+ 1|t(X′), and the transition probability h(X,X′) by the following formula
Based on the above lemma, we can recover the SIS parameters τi and μi, from h(Xt,Xt+ 1).
Let \(\xi _{i} = \prod \limits _{j:v_{j}\in \mathcal {N}_{t}(v_{i})} \left ((1-\tau _{i}){\pi _{t}^{j}}(1) + {\pi _{t}^{j}}(0) \right ) = \prod \limits _{j:v_{j}\in \mathcal {N}_{t}(v_{i})} \left (1-\tau _{i}{\pi _{t}^{j}}(1) \right ) \approx 1-\tau _{i} \cdot {\sum }_{j: v_{j}\in \mathcal {N}_{t}(v_{i})} {\pi _{t}^{j}}(1). \) Then we have
and
The proposed method provides an effective way to infer HAI parameters. Even if complete information on HAI parameters is not available initially, we can resort to this approach to improve estimation of the parameters with the updating observations and the tracked human contact networks. While theoretically the Baum-Welch reestimate converges to a local maximum, our computational experiments to be presented in “Computational study” show that our proposed approach has a good performance in the sense that the estimated values are close to the actual ones.
Computational study
In this section, we carry out a computational study, based on a real-world healthcare setting and real-world human tracking data collected from the facility, for assessing the performance of our proposed solution framework and conducting a comparative analysis with other existing epidemic models.
Baseline algorithms
We compare our proposed methods of individual-level transmission tracking approaches – detection (ILTT-DT), tracing (ILTT-TR), one-step-ahead prediction (ILTT-PD1) and pure prediction (ILTT-PD0) – with the following three baseline methods.
-
Ordinary-differential-equation-based SIS model (ODE-SIS) assumes homogeneous populations. All individuals share the same infection rate τ and recovery rate μ. The initial number of infected individuals I0 is an input. The output is the number of infected individuals as a function of time determined by \( I(t) = {I_{\infty }}/\left ({1+\nu e^{-(\tau -\mu )(t-t_{0})} }\right ) \), where ν = I∞/I0 − 1 and I∞ = (τ − μ)n/τ.
-
Percolation method (Percolation) is based on probability generating functions and considers disease spread over a heterogeneous network [14]. It requires network degree distributions as input and returns the number of infected individuals at steady state.
-
Non-linear dynamical system (NLDS) uses the probability of infection vector (pt) to approximate the infection dynamics and model the evolution of epidemic outbreak over a time-varying network [15]. pt is determined by pt+ 1 = g(pt), where the non-linear function g is defined by pi,t+ 1 = 1 − μpi,t − (1 − pi,t)ξt(i), and \( \xi _{t}(i)={\prod }_{j\in \{1,\cdots ,n\}} (1-\tau A_{t}(i,j)p_{j,t}) \). This approach requires an adjacency matrix At to represent the time-varying network at time t.
Setup of experiments
We leveraged the RFID human tracking data collected from two medical wards at PWH, which suffered from a nosocomial outbreak the 2003 SARS, for conducting our computational experiments. The data consists of time-stamped locations of 56 patients and 70 healthcare workers in two medical wards over a period of four months. Indoor locations of the tracked objects were recorded every 3 seconds with a spatial resolution of 0.5 meter. We set the time threshold ΔTth = 30 minutes and the distance threshold Dth = 1 meter. While these threshold distance and time were chosen to illustrate our idea, our algorithm allows the user to specify these threshold values. We also note that the threshold distance and time depend on the type of the HAI. We constructed static daytime networks and nighttime human contact networks for each time period based on the tracking data; a time-varying hierarchical contact network of 240 time periods was then obtained.
We considered the nosocomial outbreak of the 2013 MERS-CoV in Saudi Arabia [3] for deriving practical HAI parameters.
From April 1 to May 23, 2013, thirty-four individuals, including four healthcare workers, acquired MERS-CoV in three healthcare institutions. Fever, cough, shortness of breath and gastrointestinal symptoms were respectively observed in 20, 20, 11 and 8 individuals. As of June 12, 2013 a total of 15 deaths were related to the disease. We prioritized the observations fever, shortness of breath, gastrointestinal symptoms and cough, in a descending order of priority (from 4 to 1). If multiple observations were found at the same time period for an individual, we consider the individual is at the observation state of the highest priority. There were only two health states; each individual either is susceptible (state 0) or infected (state 1). Table 2 provides the health state-observation probability matrix. We include the observation state 0, which indicates that no symptom was found.
In the experiments, we simulated the health states and observations based on the above setup.
Marco-level phenomena of hospital outbreaks
Most existing epidemic models focus on the macro-level phenomena of epidemic thresholds and the infected population. Figure 8 shows the fraction of infections at the steady state for different values of τ/μ on three static networks of snapshots at different time periods, G1, G41 and G62, respectively. G1 and G41 have the same degree distribution whereas G62 differs from them and is more sparse. The mean degrees of three networks are 15, 15 and 4, respectively. Although G1 and G41 have the same degree distribution, the underlying networks are not equivalent. As we observe from Fig. 8a and b, the simulated threshold effects of G1 and G41 are different: the curves of G1 “take off” at around τ/μ = 0.5 while the ones of G41 “take off” at around τ/μ = 0.2. Percolation gives the same threshold, τ/μ = 0.4, for G1 and G41 because it considers only the degree distribution of a network but ignores other network properties. ODE-SIS gives the same threshold for three different networks because it assumes a homogeneous connection. NLDS captures the difference between the networks but deviates much from the simulation result. Our ILTT-PD1 predicts the thresholds to be 0.5, 0.2 and 1.2 for G1, G41 and G62, respectively, which are more consistent with the simulated results compared with the other approaches.
Figure 9 shows the plot of infection fraction at different time points resulting from the models for an HAI with infection rate τ = 0.4 and recovery rate μ = 0.3. Since Percolation is not applicable for investigating transient states over time, we compare only ILTT-PD1, NLDS and ODE-SIS with the simulated result. The plots of infection fraction resulting from ILTT-PD1, NLDS and Simulation all increase rapidly at t = 3, whereas ODE-SIS increases at a significantly lower rate. As expected, ODE-SIS provides the same prediction for three networks because it does not capture the network structures. The simulated infection fraction at steady state ranges from 0.03 to 0.26. Our ILTT-PD1 provides a similar prediction to the simulated results (from 0.03 to 0.30), while NLDS has a significantly deviated prediction ranging from 0.33 to 0.54.
Among the baseline algorithms, only NLDS applies to time-varying networks. Figure 10 shows the comparison of NLDS, ILTT-PD1, ILTT-PD0 and simulated results on the time-varying network G0:T. As shown in Fig. 10a, ILTT-PD1 predicts the “take-off” of the outbreak size at a threshold τ/μ = 0.4 while NLDS and ILTT-PD0 predict a lower threshold of 0.3. Figure 10b shows the plot of infection fractions above and below the threshold at different time points. Above the threshold the infection reaches a steady state much higher than the starting point, and below the threshold the infection decays and dies out. Note that ILTT-PD0 and NLDS give almost the same prediction in both figures. The reason is that our model reduces to NLDS when observations of HAIs are not available. Taking advantage of observable information improves the accuracy of prediction, which leads to a better performance of ILTT-PD1 than ILTT-PD0.
Individual-level transmission tracking
Our proposed method has the capability to track the transmission of HAIs at an individual level. In other words, it infers the hidden health state of any person at any time. In this subsection, we compare the identification results obtained from ILTT-PD1, ILTT-DT, ILTT-TR and NLDS with fixed infection rate τ = 0.03 and recovery rate μ = 0.02. Outcomes resulting from ILTT-PD0 are identical to NLDS. Figure 11 shows the estimation of the illness evolution of person v1, who is the patient zero of an HAI. This person stayed infected (at state 1) until time period 103 and remains susceptible (at state 0) from that time onwards. ILTT-DT, ILTT-TR, ILTT-PD1 all capture the state transition close to time step 103 whereas NLDS gives a smooth curve with no clear indication of such change in state. ILTT-TR identifies hidden states with the highest accuracy, and the ILTT-DT performs better than ILTT-PD1.
The Receiver Operating Characteristic (ROC) curves in Fig. 12 exhibit a consistent trend with the results shown in Fig. 11. We observe that ILTT-TR demonstrates an advantage over the other approaches and ILTT-DT performs slightly better than ILTT-PD1 whereas NLDS’s performance is the worst. Their difference in performance is due to the fact that the approaches utilize different degrees of observable information. ILTT-TR makes use of all available observable information to obtain the estimation of the hidden Markov processes, while NLDS utilizes no observations at all. ILTT-PD1 and ILTT-DT both use past observations, but ILTT-DT performs slightly better because it also captures observable information at present.
When an HAI outbreak takes place, the healthcare organization conducts contact tracing to identify the index case and construct epidemiological links with a manual approach. If this patient zero has had frequent contacts with the others, the transmission path can only be estimated based on experience [3, 5]. Our method provides an effective tool to construct the transmission network automatically and accurately. For example, if we consider a person as being infected if his/her tracing probability is greater than 0.5, we can draw a transmission map of the hospital outbreak for the first month, as shown in Fig. 13. 27 individuals in total got infected in the first month. The patient zero v1 transmitted the disease to 4 other individuals and patient v3 is a “super-spreader” who infected 7 people. This transmission map is similar to the one reported in [3], a real-world hospital outbreak of the MERS-CoV.
Estimation of HAI parameters. \(\overline {\tau }_{p}\) and \(\overline {\mu }_{p}\) (\(\overline {\tau }_{c}\) and \(\overline {\mu }_{c}\)) respectively denote the estimated infection and recovery rates for patient (caregiver) groups. \(\overline {\phi }_{i}\) and \(\overline {\rho }_{i}\) respectively represent the estimated observation probability matrix and state distribution
HAI parameter estimation
In general, the exact values of HAI parameters were not known exactly. Existing models, which are highly sensitive to the precision of HAI parameters, might produce predictions much deviated from the actual situation if parameters are not determined correctly. Our solution framework is capable of refining the estimate of HAI parameters in a step-by-step manner based on available information of observable features and human contacts. As discussed in “HAI parameter estimation”, our solution framework learns the infection rate τi and the recovery rate μi for any individual vi. Without loss of generality, we set τp and μp for the patient group, and τc and μc for the caregiver group. The following experiment illustrates the estimation of parameters of SIS dynamics.
Figure 14a and b show the estimation of infection rates and recovery rates for patients and caregivers. The dashed lines indicate true parameter values while the solid lines represent the estimated values at each step of estimation. In Fig. 14a, the true value of τp is 0.3 and the estimate \( \bar {\tau }_{p} \) converges to 0.32 after 6 runs using the proposed method. The true value of τc is 0.03 and \( \bar {\tau }_{c} \) reaches 0.01 after 4 runs. From Fig. 14b, we observe that \( \bar {\mu }_{p} \) and \( \bar {\mu }_{c} \) converge very quickly to their true values as well.
Figure 14c and d show the average gap between an estimate and the true value for the observation matrix and the initial state distribution. For an m × n matrix B, we define the average gap as
where \( \bar {e}_{i,j} \) is an estimate of element ei,j. In Fig. 14 (c), three sets of estimated observation probability matrices are given with initial average gaps AveGap\(_{0} (\bar {\phi }_{1})= 0.12 \) , AveGap\(_{0} (\bar {\phi }_{2})= 0.18 \) , and AveGap\(_{0} (\bar {\phi }_{3})= 0.31 \) , respectively, where \( \bar {\phi }_{3} \) is generated randomly.
Even though we start with these initial guesses with fairly large average gaps, they converge to zero after only a few iterations. In contrast, Fig. 14d shows that the performance of the inference method is sensitive to the initial guess of the initial state distribution ρ. As the initial guess deviates from the true value gradually from \( \bar {\rho }_{1} \) to \( \bar {\rho }_{3} \), the gaps between the final estimates and the true value become larger. For \( \bar {\rho }_{3} \), there are oscillations and the estimation converges rather slowly but the estimate is far away from the true value. This indicates the importance of an appropriate initial guess of the initial state distribution.
Conclusion
In this work, we propose a solution methodology integrating the techniques of network epidemiology and coupled hidden Markov models to infer the health state of any person at any time in a healthcare setting. We utilize advanced real-time positioning technologies for tracing person-to-person contacts among individuals, including patients and healthcare workers, in the healthcare facility and construct a time-varying human contact network. We also develop the algorithms for transmission tracking of individuals, with a given set of HAI parameters. We finally propose an estimation procedure to infer unknown HAI parameters to tackle the practical problem that the parameters are not completely known.
We conduct experiments based on four-month human tracking data collected from two medical wards at PWH, which suffered from the 2003 SARS nosocomial outbreak. Computational results show that our framework provides more accurate results for predicting macro-level phenomena such as the number of infected individuals and epidemic threshold, compared to existing epidemic models.
References
Magill, S. S., Edwards, J. R., Bamberg, W., Beldavs, Z. G., Dumyati, G., Kainer, M. A., Lynfield, R., Maloney, M., McAllister-Hollod, L., Nadle, J. et al., Multistate point-prevalence survey of health care–associated infections. N. Engl. J. Med. 370(13):1198–1208, 2014.
Centers for disease control and prevention. HAI data and statistics, 2016
Assiri, A., McGeer, A., Perl, T.M., Price, C.S., Al Rabeeah, A.A., AT Cummings, D., Alabdullatif, Z.N., Assad, M., Almulhim, A., Makhdoom, H., et al., Hospital outbreak of Middle East respiratory syndrome coronavirus. N. Engl. J. Med. 369(5):407–416, 2013.
Peiris, J.S.M., Chu, C.M., Cheng, V.C.C., Chan, K.S., Hung, I.F.N., Poon, L.L.M., Law, K.I., Tang, B.S.F., Hon, T.Y.W., Chan, C.S., et al., Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet 361(9371):1767–1772, 2003.
Lee, N., Hui, D., Wu, A., Chan, P., Cameron, P., Joynt, G.M., Ahuja, A., Yee Yung, M., Leung, C.B., To, K.F., et al., A major outbreak of severe acute respiratory syndrome in Hong Kong. N. Engl. J. Med. 348(20):1986–1994, 2003.
SARS expert committee of Hong Kong SAR government. The SARS epidemic, 2003
Hung, L.S., The SARS epidemic in Hong Kong: what lessons have we learned? J. R. Soc. Med. 96(8):374–378, 2003.
Cheng, C.-H., and Kuo, Y.-H., RFID Analytics for hospital ward management. Flex. Serv. Manuf. J. 28 (4):593–616, 2016.
Kermack, W.O., and McKendrick, A.G.: A contribution to the mathematical theory of epidemics. In: Proceedings of the Royal Society of London A: Mathematical, Physical and Engineering Sciences, vol. 115, pp. 700–721 The Royal Society , 1927.
Anderson, R.M, May, R.M., and Anderson, B., Infectious diseases of humans: dynamics and control. Aust. J. Public Health 16:208–212, 1992.
Hethcote, H.W., The mathematics of infectious diseases. SIAM Rev. 42(4):599–653, 2000.
Liljeros, F., Edling, C.R., Amaral, L.A.N., Stanley, E.H., and Åberg, Y., The web of human sexual contacts. Nature 411(6840):907–908, 2001.
Meyers, L., Contact network epidemiology: Bond percolation applied to infectious disease prediction and control. Bull. Am. Math. Soc. 44(1):63–86, 2007.
Newman, M.E.J., Spread of epidemic disease on networks. Phys. Rev. E 66(1):016128, 2002.
Prakash, B.A., Tong, H., Valler, N., Faloutsos, M., and Faloutsos, C.: Virus propagation on time-varying networks: Theory and immunization algorithms. In: Machine Learning and Knowledge Discovery in Databases, pp. 99–114. Springer, 2010.
Keeling, M.J., and Eames, K.T.D., Networks and epidemic models. J. R. Soc. Interface. 2(4):295–307, 2005.
Newman, M., Barabasi, A.-L., and Watts, D.J., The structure and dynamics of networks. New Jersey: Princeton University Press, 2006.
Keeling, M.J., and Rohani, P., Modeling infectious diseases in humans and animals. New Jersey: Princeton University Press, 2008.
Pastor-Satorras, R., Castellano, C., Mieghem, P.V., and Vespignani, A., Epidemic processes in complex networks. Rev. Mod. Phys. 87(3):925, 2015.
Nowzari, C., Preciado, V.M., and Pappas, G.J., Analysis and control of epidemics: A survey of spreading processes on complex networks. IEEE Control. Syst. 36(1):26–46, 2016.
Keeling, M.J., The effects of local spatial structure on epidemiological invasions. Proc. R. Soc. Lond. Ser. B Biol. Sci. 266(1421):859–867, 1999.
Eames, K.T.D., and Keeling, M.J., Modeling dynamic and network heterogeneities in the spread of sexually transmitted diseases. Proc. Natl. Acad. Sci. 99(20):13330–13335, 2002.
Meyers, L.A., Pourbohloul, B., Newman, M.E.J., Skowronski, D.M., and Brunham, R.C., Network theory and SARS: predicting outbreak diversity. J. Theor. Biol. 232(1):71–81, 2005.
Newman, M.E.J., Threshold effects for two pathogens spreading on a network. Phys. Rev. Lett. 95(10):108701, 2005.
Karrer, B., and Newman, M.E.J., Competing epidemics on complex networks. Phys. Rev. E 84(3):036106, 2011.
Volz, E., SIR dynamics in random networks with heterogeneous connectivity. J. Math. Biol. 56(3):293–310, 2008.
Larson, R.C., Simple models of influenza progression within a heterogeneous population. Oper. Res. 55(3): 399–412, 2007.
Teytelman, A., and Larson, R.C., Modeling influenza progression within a continuous-attribute heterogeneous population. Eur. J. Oper. Res. 220(1):238–250, 2012.
Yaesoubi, R., and Cohen, T., Generalized markov models of infectious disease spread: A novel framework for developing dynamic health policies. Eur. J. Oper. Res. 215(3):679–687, 2011.
Bansal, S., Grenfell, B.T., and Meyers, L.A., When individual behaviour matters: homogeneous and network models in epidemiology. J. R. Soc. Interface. 4(16):879–891, 2007.
Meyers, L.A., Newman, M.E.J., Martin, M., and Schrag, S., Applying network theory to epidemics: control measures for Mycoplasma pneumoniae outbreaks. Emerg. Infect. Dis. 9(2):204–210, 2003.
Liljeros, F., Giesecke, J., and Holme, P., The contact network of inpatients in a regional healthcare system. a longitudinal case study. Math. Popul. Stud. 14(4):269–284, 2007.
Ueno, T., and Masuda, N., Controlling nosocomial infection based on structure of hospital social networks. J. Theor. Biol. 254(3):655–666, 2008.
Curtis, D.E., Hlady, C.S., Pemmaraju, S.V., Polgreen, P.M., and Segre, A.M.: Modeling and estimating the spatial distribution of healthcare workers. In: Proceedings of the 1st ACM International Health Informatics Symposium, pages 287–296. ACM , 2010.
Dong, W., Pentl, Y., and Heller, K.A.: Graph-coupled HMMs for modeling the spread of infection. In: Proceedings of the 30th International Conference on Machine Learning. Citeseer , 2013.
Camci, F., and Chinnam, R.B., Health-state estimation and prognostics in machining processes. IEEE Trans. Autom. Sci. Eng. 7(3):581–597, 2010.
Feng, L., Wang, H., Si, X., and Zou, H., A state-space-based prognostic model for hidden and age-dependent nonlinear degradation process. IEEE Trans. Autom. Sci. Eng. 10(4):1072–1086, 2013.
Lawrence, R., A tutorial on hidden Markov models and selected applications in speech recognition. Proc. IEEE 77(2):257–286, 1989.
Kitagawa, G., Non-Gaussian state-space modeling of nonstationary time series. J. Am. Stat. Assoc. 82(400): 1032–1041, 1987.
Doucet, A., Godsill, S., and Andrieu, C., On sequential Monte Carlo sampling methods for Bayesian filtering. Stat. Comput. 10(3):197–208, 2000.
Baum, L.E., Eagon, J.A., et al., An inequality with applications to statistical estimation for probabilistic functions of Markov processes and to a model for ecology. Bull. Am. Math. Soc. 73(3):360–363, 1967.
Baum, L.E., Petrie, T., Soules, G., and Weiss, N., A maximization technique occurring in the statistical analysis of probabilistic functions of Markov chains. Ann. Math. Stat.,164–171, 1970.
Acknowledgments
We would like to thank patients and staff of Ward 11A and 11C of Prince of Wales Hospital for taking part in this study. Further we express our gratitude to Research Grants Council (RGC) of Hong Kong for providing financial support through GRF to this work (Project No. 14201314 and 14209416). The research of the second author was partially supported by RGC GRF (Project No. 14202115) and Health and Medical Research Fund, Food and Health Bureau, the Hong Kong SAR Government (Project No. 14151771).
Funding
This study was funded by Research Grants Council (RGC) of Hong Kong (Project No. 14201314 and 14209416). The research of the second author was partially supported by RGC GRF (Project No. 14202115) and Health and Medical Research Fund, Food and Health Bureau, the Hong Kong SAR Government (Project No. 14151771).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interests
CH Cheng declares that he has no conflict of interest. YH Kuo declares that he has no conflict of interest. Z Zhou declares that he has no conflict of interest.
Ethical Approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. This article does not contain any studies with animals performed by any of the authors.
Informed Consent
Informed consent was obtained from all individual participants included in the study.
Additional information
This article is part of the Topical Collection on Mobile & Wireless Health
Appendix: Proofs
Appendix: Proofs
Theorem 1
The solution to the problem of individual-level detection is given by
and the basic solution to the detection problem can be factored in a product form
Proof
By the definition of the one-step-ahead prediction probability, stated in Eq. 7, and the independence assumption, stated in Eq. 8, we have
The denominator of the basic solution (5) can be rewritten as
The third equality follows from the Markov property. In coupled HMMs, observations are independent of each other and an observation is only affected by its own state. Thus we have
By Eqs. 5, 9, 23, 24 and 25, we have \( \pi _{t} = \prod \nolimits _{i = 1} {\pi _{t}^{i}} \) and \( {\pi _{t}^{i}} = \frac {{\phi _{t}^{i}} \pi _{t|t-1}^{i}}{\sum \nolimits _{{x_{t}^{i}}} {\phi _{t}^{i}} \pi _{t|t-1}^{i}} \). □
Theorem 2
The solution to the problem of individual-level one-step-ahead prediction is given by
where \( p_{t|t-1}^{i} = {\prod }_{j:v_{j} \in \mathcal {N}_{t-1} (v_{i})} \left (\pi _{t-1}^{j} (1) \!\cdot \! (1 - \tau _{i}) + \pi _{t-1}^{j}(0) \right ) \).
Proof
\(\pi _{t|t-1}^{i} (0)\) is the estimate that person vi is not infected at time t based on the information about the person-to-person contact history and features of all individuals by time t − 1. We have
According to the SIS dynamics, if \( x_{t-1}^{i}= 1 \), \( {x_{t}^{i}} \) is independent of \( x_{t-1}^{j} \) for all j≠i. That is,
If \( x_{t-1}^{i}= 0 \), \( {x_{t}^{i}} \) is influenced by other \( x_{t-1}^{j}, j \neq i \). Let \( X_{t-1}^{\sim i} \) be a state vector excluding \( x_{t-1}^{i} \), or formally, \( X_{t-1}^{\sim i} = (x_{t-1}^{1},x_{t-1}^{2},..., x_{t-1}^{i-1},x_{t-1}^{i + 1},...,x_{t-1}^{n})\). Let
We apply the mean-field analysis approach to decompose the effect of \( X_{t-1}^{\sim i} \) on \( {x_{t}^{i}} \) into single average effects. A neighbor vj of vi is infected at time t − 1 with probability \( \pi _{t-1}^{j}(1) \), and is susceptible with probability \( \pi _{t-1}^{j}(0) \). The average effect of vj on vi for which vi stays susceptible is \( \pi _{t-1}^{j}(1) \cdot (1-\tau _{i}) + \pi _{t-1}^{j}(0) \cdot 1 \). Thus we have
and this completes the proof. □
Theorem 3
The solution to the problem of individual-level tracing is provided by
and the basic solution to the tracing problem can be factored in the following product form:
where s ≤ t, \( P(x_{s + 1}^{i}= 0|{\pi _{s}^{i}}= 1)=\mu _{i} \), and \( P(x_{s + 1}^{i}= 1|{\pi _{s}^{i}}= 1)= 1-\mu _{i} \).
Proof
We first prove the claim that \( \pi _{s|t} = \prod \nolimits _{i} \pi _{s|t}^{i} \) by backward induction. When s = t, this equality holds since \( \pi _{s|t} = \pi _{t} = \prod \nolimits _{i} {\pi _{t}^{i}} \). Now suppose \( \pi _{s + 1|t} = {\prod }_{i} \pi _{s + 1|t}^{i} \) for s ≤ t − 1. We have
The second equality holds because Xs+ 1 is a combination of \( x_{s + 1}^{i} \). From the above equation and the tracing formula, we have \( \pi _{s|t} = \prod \nolimits _{i} \pi _{s|t}^{i} \) for s ≤ t − 1, and the claim holds.
As \( h_{s + 1|s}^{i} = P(x_{s + 1}^{i}|X_{s}) \), we aim to decompose the coupling variable Xs. Note that an infected person is not affected by other individuals according to SIS dynamics. Thus we only need to consider the case of \( {x_{s}^{i}}= 1 \). We have
and it completes the proof. □
Lemma 1
The probability πt,t+ 1|T(X,X′) can be derived from the detection probability πt(X), the tracing probability πt+ 1|T(X′), the one-step-ahead prediction probability πt+ 1|t(X′), and the transition probability h(X,X′) by the following formula
Proof
Let αt and βt be the forward and backward variables respectively as in standard HMMs. Note that
By the above relationships, we have
□
Rights and permissions
About this article
Cite this article
Cheng, CH., Kuo, YH. & Zhou, Z. Tracking Nosocomial Diseases at Individual Level with a Real-Time Indoor Positioning System. J Med Syst 42, 222 (2018). https://doi.org/10.1007/s10916-018-1085-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10916-018-1085-4