Abstract
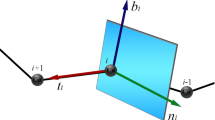
Parvalbumin (Parv) is a typical protein with EF-hand motifs that play an important role in many physiological processes. We present a novel free energy to model the skeletal C\(_\alpha \) chain of the protein from the basic principle of mathematics and physics. Starting from the crystal structure of Parv (PDB code 2PVB), we first analyze the profile of the C\(_\alpha \) bond and torsion angles over the segment that contains the secondary structures. Then the parameters in the energy function are evaluated for the helix ABCD fragment that contains two EF-hand domains in Parv. Meanwhile an eight-soliton configuration at the energy minimum is constructed to model the conformation of ABCD fragment. The deviation of the conformation constructed from the model away from the crystal structure is as small as 1.28 Å. The structural modeling stems from the physical energy, which is a benefit relative to the statistics-based or knowledge-based technologies.






Similar content being viewed by others
References
R.H. Kretsinger, D.J. Nelson, Calcium in biological systems. Coord. Chem. Rev. 18, 29–124 (1976)
R.H. Kretsinger, D. Moncrief, A. Persechini, The EF-hand family of calcium-modulated proteins. Trends Neurosci. 12, 462–467 (1989)
H. Kawasaki, R.H. Kretsinger, Calcium-binding proteins 1: EF-hands. Protein Profile 1, 343–517 (1994)
B.W. Schafer, C.W. Heizmann, The S100 family of EF-hand calcium-binding proteins: functions and pathology. Trends Biochem. Sci. 21, 134–140 (1996)
A.S. Polans, D. Witkowska, T.L. Heley, D. Amundson, L. Baizer, G. Adamus, Recoverin, a photoreceptor-specific calcium-binding protein, is expressed by the tumor of a patient with cancer-associated retinopathy. Proc. Natl. Acad. Sci. U. S. A. 92, 9176–9180 (1995)
P. Vito, E. Lacana, L.D. Adamio, Interfering with apoptosis: Ca\(^{2+}\)-binding protein ALG-2 and Alzheimer’s disease gene ALG-3. Science 271, 521–525 (1996)
R.H. Kretsinger, C.E. Nockolds, Carp muscle calcium-binding protein. J. Biol. Chem. 248, 3313–3326 (1973)
S.J. Opella, D.J. Nelson, O. Jardetzky, Carbon magnetic resonance study of the conformational changes in carp muscle calcium binding parvalbumin. J. Am. Chem. Soc. 96, 7157–7159 (1974)
A. Cavé, C.M. Dobson, J. Parello, R.J.P. Williams, Conformation mobility within the structure of muscular parvalbumins. An NMR study of the aromatic resonances of phenylalanine residues. FEBS Lett. 65, 190–194 (1976)
J.P. Declecq, B. Tinant, J. Parello, X-ray structure of a new crystal form of pike 4.10 \(\beta \) parvalbumin. Acta Crystallogr. Sect. D 52, 165–169 (1996)
J.P. Declercq, C. Evrard, V. Lamzin, J. Parello, Crystal structure of the EF-hand parvalbumin at atomic resolution (0.91 Å) and at low temperature (100 K). Evidence for conformational multistates within the hydrophobic core. Protein Sci. 8, 2194–2204 (1999)
R.C. Richardson, N.M. King, D.J. Harrington, H. Sun, W.E. Royer, D.J. Nelson, X-ray crystal structure and molecular dynamics simulations of silver hake parvalbumin (isoform B). Protein Sci. 9, 73–82 (2000)
S.K. Drake, K.L. Lee, J.J. Falke, Tuning the equilibrium ion affinity and selectivity of the EF-hand calcium binding motif: substitutions at the gateway position. Biochemistry 35, 6697–6705 (1996)
K. Fahie, R. Pitts, K.M. Elkins, D.J. Nelson, Molecular dynamics study of Ca\(^{2+}\)-binding loop variants of silver hake parvalbumin with aspartic acid at the “Gateway” position. J. Biomol. Struct. Dyn. 19, 821–837 (2002)
D. Baker, A. Sali, Protein structure prediction and structural genomics. Science 294, 93–96 (2001)
K.A. Dill, S.B. Ozkan, M.S. shell, T.R. Weikl, The protein folding problem. Annu. Rev. Biophys. 37, 289–316 (2008)
P. Ahlstöm, O. Teleman, B. Jönsson, Molecular dynamics simulation of interfacial water structure and dynamics in a parvalbumin solution. J. Am. Chem. Soc. 110, 4198–4203 (1988)
D. Allouche, J. Parello, Y.H. Sanejouand, Ca\(^{2+}\)/Mg\(^{2+}\) exchange in parvalbumin and other EF-hand proteins. A theoretical study. J. Mol. Biol. 285, 857–873 (1999)
M.S. Cates, M.L. Teodoro, G.N. Phillops Jr., Molecular mechanisms of calcium and magnesium binding to parvalbumin. Biophys. J. 82, 1133–1146 (2002)
A.N. Kucharski, C.E. Scott, J.P. Davis, P.M. Kekenes-Huskey, Understanding ion binding affinity and selectivity in \(\beta \)-parvalbumin unsing molecular dynamics and mean spherical approximation theory. J. Phys. Chem. B 120, 8617–8630 (2016)
J.F. He, J. Dai, J. Li, X.B. Peng, A.J. Niemi, Aspects of structural landscape of human islet amyloid polypeptide. J. Chem. Phys. 142, 045102 (2015)
J. Dai, A.J. Niemi, J.F. He, A. Sieradzan, N. Ilieva, Bloch spin waves and emergent structure in protein folding with HIV envelope glycoprotein as an example. Phys. Rev. E 93, 032409 (2016)
J. Dai, A.J. Niemi, J.F. He, Conformational landscape of an amyloid intra-cellular domain and Landau–Ginzburg–Wilson paradigm in protein dynamics. J. Chem. Phys. 145, 045103 (2016)
J.J. Liu, J. Dai, J.F. He, A.J. Niemi, N. Ilieva, Multistage modeling of protein dynamics with monomeric Myc oncoprotein as an example. Phys. Rev. E 95, 032406 (2017)
X. Peng, A. Chenani, S. Hu, Y. Zhou, A. Niemi, A three dimensional visualisation approach to protein heavy-atom structure reconstruction. BMC Struct. Biol. 14, 27 (2014)
S.B. Anfinsen, Principles that govern the folding of protein chains. Science 181, 223–230 (1973)
I. Sillitoe, T.E. Lewis, A. Cuff, S. Das, P. Ashford, N.L. Dawson, N. Furnham, R.A. Laskowski, D. Lee, J.G. Lees, S. Lehtinen, R.A. Studer, J. Thornton, C.A. Orengo, CATH: comprehensive structural and functional annotations for genome sequences. Nucleic Acids Res. 43(D1), D376–D381 (2015)
A.G. Murzin, S.E. Brenner, T. Hubbard, C. Chothia, SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 247, 536–540 (1995)
Acknowledgements
We would like to thank Prof. Antti J. Niemi of Uppsala University, our cooperator in biophysics, for continued discussions on the theory and method. We also thanks the support of the international cooperation project of Beijing Institute of Technology.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhao, Y., He, J. & Li, J. The structural modeling of EF-hand motifs in parvalbumin. J Math Chem 56, 2525–2536 (2018). https://doi.org/10.1007/s10910-018-0904-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10910-018-0904-7