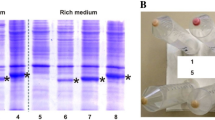
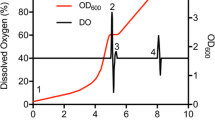
Abstract
The isomerization of a covalently bound retinal is an integral part of both microbial and animal rhodopsin function. As such, detailed structure and conformational changes in the retinal binding pocket are of significant interest and are studied in various NMR, FTIR, and Raman spectroscopy experiments, which commonly require isotopic labeling of retinal. Unfortunately, the de novo organic synthesis of an isotopically-labeled retinal is complex and often cost-prohibitive, especially for large scale expression required for solid-state NMR. We present the novel protocol for biosynthetic production of an isotopically labeled retinal ligand concurrently with an apoprotein in E. coli as a cost-effective alternative to the de novo organic synthesis. Previously, the biosynthesis of a retinal precursor, β-carotene, has been introduced into many different organisms. We extended this system to the prototrophic E. coli expression strain BL21 in conjunction with the inducible expression of a β-dioxygenase and proteo-opsin. To demonstrate the applicability of this system, we were able to assign several new carbon resonances for proteorhodopsin-bound retinal by using fully 13C-labeled glucose as the sole carbon source. Furthermore, we demonstrated that this biosynthetically produced retinal can be extracted from E. coli cells by applying a hydrophobic solvent layer to the growth medium and reconstituted into an externally produced opsin of any desired labeling pattern.





Similar content being viewed by others
References
Ahuja S, Hornak V, Yan EC, Syrett N, Goncalves JA, Hirshfeld A, Ziliox M, Sakmar TP, Sheves M, Reeves PJ (2009) Helix movement is coupled to displacement of the second extracellular loop in rhodopsin activation. Nat Struct Mol Biol 16:168–175
Becker-Baldus J, Bamann C, Saxena K, Gustmann H, Brown LJ, Brown RC, Reiter C, Bamberg E, Wachtveitl J, Schwalbe H, Glaubitz C (2015) Enlightening the photoactive site of channelrhodopsin-2 by DNP-enhanced solid-state NMR spectroscopy. Proc Natl Acad Sci USA 112:9896–9901
Béja O, Aravind L, Koonin EV, Suzuki MT, Hadd A, Nguyen LP, Jovanovich SB, Gates CM, Feldman RA, Spudich JL (2000) Bacterial rhodopsin: evidence for a new type of phototrophy in the sea. Science 289:1902–1906
Béja O, Spudich EN, Spudich JL, Leclerc M, DeLong EF (2001) Proteorhodopsin phototrophy in the ocean. Nature 411:786–789
Castellani F, van Rossum B, Diehl A, Schubert M, Rehbein K, Oschkinat H (2002) Structure of a protein determined by solid-state magic-angle-spinning NMR spectroscopy. Nature 420:98–102
Creemers AF, Lugtenburg J (2002) The preparation of all-trans uniformly 13C-labeled retinal via a modular total organic synthetic strategy. Emerging central contribution of organic synthesis toward the structure and function study with atomic resolution in protein research. J Am Chem Soc 124:6324–6334
Creemers AFL, Kiihne S, Bovee-Geurts PHM, DeGrip WJ, Lugtenburg J, de Groot HJM (2002) H-1 and C-13 MAS NMR evidence for pronounced ligand-protein interactions involving the ionone ring of the retinylidene chromophore in rhodopsin. Proc Natl Acad Sci USA 99:9101–9106
Cunningham FX Jr, Gantt E (2005) A study in scarlet: enzymes of ketocarotenoid biosynthesis in the flowers of Adonis aestivalis. Plant J 41:478–492
Dawadi P, Lugtenburg J (2010) Synthesis and use of stable isotope enriched retinals in the field of vitamin A. Molecules 15:1825–1872
Dioumaev AK, Brown LS, Shih J, Spudich EN, Spudich JL, Lanyi JK (2002) Proton transfers in the photochemical reaction cycle of proteorhodopsin. Biochemistry 41:5348–5358
Ernst OP, Lodowski DT, Elstner M, Hegemann P, Brown LS, Kandori H (2014) Microbial and animal rhodopsins: structures, functions, and molecular mechanisms. Chem Rev 114:126–163
Friedrich T, Geibel S, Kalmbach R, Chizhov I, Ataka K, Heberle J, Engelhard M, Bamberg E (2002) Proteorhodopsin is a light-driven proton pump with variable vectoriality. J Mol Biol 321:821–838
Fung BM, Khitrin AK, Ermolaev K (2000) An improved broadband decoupling sequence for liquid crystals and solids. J Magn Reson 142:97–101
Govorunova EG, Sineshchekov OA, Li H, Spudich JL (2017) Microbial rhodopsins: diversity, mechanisms, and optogenetic applications. Annu Rev Biochem 86:845–872
Harbison G, Mulder P, Pardoen H, Lugtenburg J, Herzfeld J, Griffin R (1985a) High-resolution carbon-13 NMR of retinal derivatives in the solid state. J Am Chem Soc 107:4809–4816
Harbison GS, Smith SO, Pardoen JA, Courtin JM, Lugtenburg J, Herzfeld J, Mathies RA, Griffin RG (1985b) Solid-state carbon-13 NMR detection of a perturbed 6-s-trans chromophore in bacteriorhodopsin. Biochemistry 24:6955–6962
Hartmann SR, Hahn EL (1962) Nuclear double resonance in the rotating frame. Phys Rev 128:2042–2053
Jang H-J, Yoon S-H, Ryu H-K, Kim J-H, Wang C-L, Kim J-Y, Oh D-K, Kim S-W (2011) Retinoid production using metabolically engineered Escherichia coli with a two-phase culture system. Microb Cell Fact 10:1–12
Jang HJ, Ha BK, Kim JW, Jung KH, Ahn J, Yoon SH, Kim SW (2014) Comparison of extraction phases for a two-phase culture of a recombinant E. coli producing retinoids. Biotechnol Lett 36:497–505
Kaur J, Kriebel CN, Eberhardt P, Jakdetchai O, Leeder AJ, Weber I, Brown LJ, Brown RCD, Becker-Baldus J, Bamann C, Wachtveitl J, Glaubitz C (2018) Solid-state NMR analysis of the sodium pump Krokinobacter rhodopsin 2 and its H30A mutant. J Struct Biol. https://doi.org/10.1016/j.jsb.2018.06.001
Kim SY, Waschuk SA, Brown LS, Jung K-H (2008) Screening and characterization of proteorhodopsin color-tuning mutations in Escherichia coli with endogenous retinal synthesis. Biochim Biophys Acta 1777:504–513
Klare JP, Chizhov I, Engelhard M (2008) Microbial rhodopsins: scaffolds for ion pumps, channels, and sensors. Results Probl Cell Differ 45:73–122
Krebs RA, Dunmire D, Partha R, Braiman MS (2003) Resonance Raman characterization of proteorhodopsin’s chromophore environment. J Phys Chem B 107:7877–7883
Lansing JC, Hohwy M, Jaroniec CP, Creemers A, Lugtenburg J, Herzfeld J, Griffin RG (2002) Chromophore distortions in the bacteriorhodopsin photocycle: evolution of the H-C14-C15-H dihedral angle measured by solid-state NMR. Biochemistry 41:431–438
Leeder AJ, Brown LJ, Becker-Baldus J, Mehler M, Glaubitz C, Brown RCD (2018) Synthesis of isotopically labeled all-trans retinals for DNP-enhanced solid-state NMR studies of retinylidene proteins. J Label Comp Radiopharm 61:922–933
LeMaster DM, Kushlan DM (1996) Dynamical mapping of E-coli thioredoxin via C-13 NMR relaxation analysis. J Am Chem Soc 118:9255–9264
Lugtenburg J, Creemers AFL, Verhoeven MA, van Wijk AAC, Verdegem PJE, Monnee MCF, Jansen FJHM (1999) Synthesis of C-13-labeled carotenoids and retinoids. Pure Appl Chem 71:2245–2251
Man D, Wang W, Sabehi G, Aravind L, Post AF, Massana R, Spudich EN, Spudich JL, Béjà O (2003) Diversification and spectral tuning in marine proteorhodopsins. EMBO J 22:1725–1731
Mao J, Do NN, Scholz F, Reggie L, Mehler M, Lakatos A, Ong YS, Ullrich SJ, Brown LJ, Brown RC, Becker-Baldus J, Wachtveitl J, Glaubitz C (2014) Structural basis of the green-blue color switching in proteorhodopsin as determined by NMR spectroscopy. J Am Chem Soc 136:17578–17590
Mehler M, Scholz F, Ullrich SJ, Mao J, Braun M, Brown LJ, Brown RC, Fiedler SA, Becker-Baldus J, Wachtveitl J (2013) The EF loop in green proteorhodopsin affects conformation and photocycle dynamics. Biophys J 105:385–397
Mehler M, Eckert CE, Leeder AJ, Kaur J, Fischer T, Kubatova N, Brown LJ, Brown RCD, Becker-Baldus J, Wachtveitl J, Glaubitz C (2017) Chromophore distortions in photointermediates of proteorhodopsin visualized by dynamic nuclear polarization-enhanced solid-state NMR. J Am Chem Soc 139:16143–16153
Misawa N, Nakagawa M, Kobayashi K, Yamano S, Izawa Y, Nakamura K, Harashima K (1990) Elucidation of the Erwinia uredovora carotenoid biosynthetic pathway by functional analysis of gene products expressed in Escherichia coli. J Bacteriol 172:6704–6712
Morcombe CR, Zilm KW (2003) Chemical shift referencing in MAS solid state NMR. J Magn Reson 162:479–486
Morcombe CR, Gaponenko V, Byrd RA, Zilm KW (2004) Diluting abundant spins by isotope edited radio frequency field assisted diffusion. J Am Chem Soc 126:7196–7197
Paine JA, Shipton CA, Chaggar S, Howells RM, Kennedy MJ, Vernon G, Wright SY, Hinchliffe E, Adams JL, Silverstone AL (2005) Improving the nutritional value of Golden Rice through increased pro-vitamin A content. Nat Biotechnol 23:482–487
Pfleger N, Lorch M, Woerner AC, Shastri S, Glaubitz C (2008) Characterisation of Schiff base and chromophore in green proteorhodopsin by solid-state NMR. J biomol NMR 40:15–21
Pines A, Gibby MG, Waugh JS (1972) Proton-enhanced nuclear induction spectroscopy. A method for high resolution NMR of dilute spins in solids. J Chem Phys 56:1776–1777
Ran T, Ozorowski G, Gao Y, Sineshchekov OA, Wang W, Spudich JL, Luecke H (2013) Cross-protomer interaction with the photoactive site in oligomeric proteorhodopsin complexes. Acta Crystallogr D Biol Crystallogr 69:1965–1980
Reckel S, Gottstein D, Stehle J, Löhr F, Verhoefen MK, Takeda M, Silvers R, Kainosho M, Glaubitz C, Wachtveitl J (2011) Solution NMR structure of proteorhodopsin. Angew Chem Int Ed 50:11942–11946
Rehorek M, Heyn MP (1979) Binding of all-trans-retinal to the purple membrane. Evidence for cooperativity and determination of the extinction coefficient. Biochemistry 18:4977–4983
Rohmer M, Knani M, Simonin P, Sutter B, Sahm H (1993) Isoprenoid biosynthesis in bacteria: a novel pathway for the early steps leading to isopentenyl diphosphate. Biochem J 295:517–524
Sabehi G, Loy A, Jung K-H, Partha R, Spudich JL, Isaacson T, Hirschberg J, Wagner M, Béjà O (2005) New insights into metabolic properties of marine bacteria encoding proteorhodopsins. PLoS Biol 3:e273
Shi L, Ahmed MA, Zhang W, Whited G, Brown LS, Ladizhansky V (2009a) Three-dimensional solid-state NMR study of a seven-helical integral membrane proton pump—structural insights. J Mol Biol 386:1078–1093
Shi L, Lake EM, Ahmed MA, Brown LS, Ladizhansky V (2009b) Solid-state NMR study of proteorhodopsin in the lipid environment: secondary structure and dynamics. Biochim Biophys Acta 1788:2563–2574
Shi L, Kawamura I, Jung KH, Brown LS, Ladizhansky V (2011) Conformation of a seven-helical transmembrane photosensor in the lipid environment. Angew Chem Int Ed Engl 50:1302–1305
Smith SO, Lugtenburg J, Mathies RA (1985) Determination of retinal chromophore structure in bacteriorhodopsin with resonance Raman spectroscopy. J Membr Biol 85:95–109
Smith SO, De Groot HJ, Gebhard R, Courtin JM, Lugtenburg J, Herzfeld J, Griffin RG (1989) Structure and protein environment of the retinal chromophore in light-and dark-adapted bacteriorhodopsin studied by solid-state NMR. Biochemistry 28:8897–8904
Spudich JL, Yang C-S, Jung K-H, Spudich EN (2000) Retinylidene proteins: structures and functions from archaea to humans. Annu Rev Cell Dev Biol 16:365–392
Takegoshi K, Nakamura S, Terao T (2001) C-13-H-1 dipolar-assisted rotational resonance in magic-angle spinning NMR. Chem Phys Lett 344:631–637
Terakita A (2005) The opsins. Genome Biol 6:213
Tomonaga Y, Hidaka T, Kawamura I, Nishio T, Ohsawa K, Okitsu T, Wada A, Sudo Y, Kamo N, Ramamoorthy A, Naito A (2011) An active photoreceptor intermediate revealed by in situ photoirradiated solid-state NMR spectroscopy. Biophys J 101:L50–L52
Verhoeven MA, Creemers AFL, Bovee-Geurts PHM, De Grip WJ, Lugtenburg J, de Groot HJM (2001) Ultra-high-field MAS NMR assay of a multispin labeled ligand bound to its G-protein receptor target in the natural membrane environment: electronic structure of the retinylidene chromophore in rhodopsin. Biochemistry 40:3282–3288
Von Lintig J, Vogt K (2000) Filling the gap in vitamin a research molecular identification of an enzyme cleaving β-carotene to retinal. J Biol Chem 275:11915–11920
Wang S, Munro RA, Shi L, Kawamura I, Okitsu T, Wada A, Kim SY, Jung KH, Brown LS, Ladizhansky V (2013a) Solid-state NMR spectroscopy structure determination of a lipid-embedded heptahelical membrane protein. Nat Methods 10:1007–1012
Wang S, Shi L, Okitsu T, Wada A, Brown LS, Ladizhansky V (2013b) Solid-state NMR (13)C and (15)N resonance assignments of a seven-transmembrane helical protein anabaena sensory rhodopsin. Biomol NMR Assign 7:253–256
Yang J, Guo L (2014) Biosynthesis of beta-carotene in engineered E. coli using the MEP and MVA pathways. Microb Cell Fact 13:160
Acknowledgements
This research was supported by Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery Grant No. RGPIN-2014-04547 to V.L. and Grant No. RGPIN-2018-04397 to L.S.B, and by Basic Science Research Program through the National Research Foundation of Korea (NRF) to K.-H.J., funded by the Ministry of Education (Grant No. 2018R1A6A1A03024940). Additionally, R.A.M. and M.E.W. were recipients of NSERC doctoral scholarship, and J.d.V. was a recipient of Ontario Graduate Scholarship (OGS). We want to thank Alicia Cronin for her help with the initial expression system testing.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Munro, R.A., de Vlugt, J., Ward, M.E. et al. Biosynthetic production of fully carbon-13 labeled retinal in E. coli for structural and functional studies of rhodopsins. J Biomol NMR 73, 49–58 (2019). https://doi.org/10.1007/s10858-019-00225-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-019-00225-9