Abstract
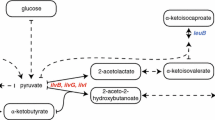
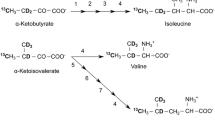
We recently developed a practical protocol for preparing proteins bearing stereo-selectively 13C-methyl labeled leucines and valines, instead of the commonly used 13C-methyl labeled precursors for these amino acids, by E. coli cellular expression. Using this protocol, proteins with any combinations of isotope-labeled or unlabeled Leu and Val residues were prepared, including some that could not be prepared by the precursor methods. However, there is still room for improvement in the labeling efficiencies for Val residues, using the methods with labeled precursors or Val itself. This is due to the fact that the biosynthesis of Val could not be sufficiently suppressed, even by the addition of large amounts of Val or its precursors. In this study, we completely solved this problem by using a mutant strain derived from E. coli BL21(DE3), in which the metabolic pathways depending on two enzymes, dihydroxy acid dehydratase and β-isopropylmalate dehydrogenase, are completely aborted by deleting the ilvD and leuB genes, which respectively encode these enzymes. The ΔilvD E. coli mutant terminates the conversion from α,β-dihydroxyisovalerate to α-ketoisovalerate, and the conversion from α,β-dihydroxy-α-methylvalerate to α-keto-β-methylvalerate, which produce the preceding precursors for Val and Ile, respectively. By the further deletion of the leuB gene, the conversion from Val to Leu was also fully terminated. Taking advantage of the double-deletion mutant, ΔilvDΔleuB E. coli BL21(DE3), an efficient and residue-selective labeling method with various isotope-labeled Ile, Leu, and Val residues was established.




Similar content being viewed by others
References
Ayala I, Hamelin O, Amero C, Pessey O, Plevin MJ, Gans P, Boisbouvier J (2012) An optimized isotopic labelling strategy of isoleucine-γ2 methyl groups for solution NMR studies of high molecular weight proteins. Chem Commun 48:1434–1436
Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H (2006) Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2006(2):0008
Datsenko KA, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA 97(12):6640–6645
Ely B, Pittard J (1979) Aromatic amino acid biosynthesis: Regulation of shikimate kinase in Escherichia coli K-12. J Bacteriol 138(3):933–943
Gans P, Hamelin O, Sounier R, Ayala I, Durá MA, Amero CD, Noirclerc-Savoye M, Franzetti B, Plevin MJ, Boisbouvier J (2010) Stereospecific isotopic labeling of methyl groups for NMR spectroscopic studies of high-molecular-weight proteins. Angew Chem Int Ed Engl 49(11):1958–1962
Gardner KH, Rosen MK, Kay LE (1997) Global folds of highly deuterated, methyl-protonated proteins by multidimensional NMR. Biochemistry 36(6):1389–1401
Goto NK, Gardner KH, Mueller GA, Willis RC, Kay LE (1999) A robust and cost-effective method for the production of Val, Leu, Ile (delta 1) methyl-protonated 15N-, 13C-, 2H-labeled proteins. J Biomol NMR 13(4):369–374
Gross JD, Gelev VM, Wagner G (2003) A sensitive and robust method for obtaining intermolecular NOEs between side chains in large protein complexes. J Biomol NMR 25(3):235–242
Hajduk PJ, Augeri DJ, Mack J, Mendoza R, Yang J, Betz SF, Fesik SW (2000) NMR-based screening of proteins containing 13C-labeled methyl groups. J Am Chem Soc 122:7898–7904
Howard BR, Endrizzi JA, Remington SJ (2000) Crystal structure of Escherichia coli malate synthase G complexed with magnesium and glyoxylate at 2.0 Å resolution: mechanistic implications. Biochemistry 39(11):3156–3168
Kainosho M, Güntert P (2009) SAIL–stereo-array isotope labeling. Q Rev Biophys 42(4):247–300
Kainosho M, Torizawa T, Iwashita Y, Terauchi T (2006) Mei Ono, A.; Güntert, P., Optimal isotope labelling for NMR protein structure determinations. Nature 440(7080):52–57
Kerfah R, Plevin MJ, Sounier R, Gans P, Boisbouvier J (2015a) Methyl-specific isotopic labeling: a molecular tool box for solution NMR studies of large proteins. Curr Opin Struct Biol 32:113–122
Kerfah R, Plevin MJ, Pessey O, Hamelin O, Gans P, Boisbouvier J (2015b) Scrambling free combinatorial labeling of alanine-β, isoleucine-δ1, leucine-proS and valine-proS methyl groups for the detection of long range NOEs. J Biomol NMR 61(1):73–82
Lichtenecker R, Ludwiczek ML, Schmid W, Konrat R (2004) Simplification of protein NOESY spectra using bioorganic precursor synthesis and NMR spectral editing. J Am Chem Soc 126(17):5348–5349
Lichtenecker RJ, Weinhäupl K, Reuther L, Schörghuber J, Schmid W, Konrat R (2013) Independent valine and leucine isotope labeling in Escherichia coli protein overexpression systems. J Biomol NMR 57(3):205–209
Lin MT, Sperling LJ, Frerick Schmidt HL, Tang M, Samoilova RI, Kumasaka T, Iwasaki T, Dikanov SA, Rienstra CM, Gennis RB (2011) A rapid and robust method for selective isotope labeling of proteins. Methods 55(4):370–378
Mas G, Crublet E, Hamelin O, Gans P, Boisbouvier J (2013) Specific labeling and assignment strategies of valine methyl groups for NMR studies of high molecular weight proteins. J Biomol NMR 57(3):251–262
Miyanoiri Y, Takeda M, Jee J, Ono AM, Okuma K, Terauchi T, Kainosho M (2011) Alternative SAIL-Trp for robust aromatic signal assignment and determination of the χ(2) conformation by intra-residue NOEs. J Biomol NMR 51(4):425–435
Miyanoiri Y, Takeda M, Okuma K, Ono AM, Terauchi T, Kainosho M (2013) Differential isotope-labeling for Leu and Val residues in a protein by E. coli cellular expression using stereo-specifically methyl labeled amino acids. J Biomol NMR 57(3):237–249
Plevin M, Boisbouvier J (2012) Isotope-labelling of methyl groups for NMR studies of large proteins. In: Clore M, Potts J (eds) Recent developments in biomolecular NMR. Royal Society of Chemistry, London, pp 1–24
Rajesh S, Nietlispach D, Nakayama H, Takio K, Laue ED, Shibata T, Ito Y (2003) A novel method for the biosynthesis of deuterated proteins with selective protonation at the aromatic rings of Phe, Tyr and Trp. J Biomol NMR 27:81–86
Ruschak AM, Kay LE (2012) Proteasome allostery as a population shift between interchanging conformers. Proc Natl Acad Sci USA 109:E3454–E3462
Ruschak AM, Velyvis A, Kay LE (2010) A simple strategy for 13C, 1H labeling at the Ile-γ2 methyl position in highly deuterated proteins. J Biomol NMR 48(3):129–135
Takeda M, Kainosho M (2012) Cell-free protein synthesis using E. coli cell extract for NMR studies. Adv Exp Med Biol 992:167–177
Takeda M, Ikeya T, Güntert P, Kainosho M (2007) Automated structure determination of proteins with the SAIL-FLYA NMR method. Nat Protoc 2(11):2896–2902
Takeda M, Hallenga K, Shigezane M, Waelchli M, Löhr F, Markley JL, Kainosho M (2011) Construction and performance of an NMR tube with a sample cavity formed within magnetic susceptibility-matched glass. J Magn Reson 209(2):167–173
Torizawa T, Shimizu M, Taoka M, Miyano H, Kainosho M (2004) Efficient production of isotopically labeled proteins by cell-free synthesis: a practical protocol. J Biomol NMR 30(3):311–325
Tugarinov V, Kay LE (2003) Ile, Leu, and Val methyl assignments of the 723-residue malate synthase G using a new labeling strategy and novel NMR methods. J Am Chem Soc 125(45):13868–13878
Tugarinov V, Kay LE (2004) 1H, 13C-1H, 1H dipolar cross-correlated spin relaxation in methyl groups. J Biomol NMR 29(3):369–376
Tugarinov V, Muhandiram R, Ayed A, Kay LE (2002) Four-dimensional NMR spectroscopy of a 723-residue protein: chemical shift assignments and secondary structure of malate synthase g. J Am Chem Soc 124(34):10025–10035
Vaiphei ST, Mao L, Shimazu T, Park JH, Inouye M (2010) Use of amino acids as inducers for high-level protein expression in the single-protein production system. Appl Environ Microbiol 76(18):6063–6068
Acknowledgments
This research was supported by the Platform Project for Supporting in Drug Discovery and Life Science Research from the Ministry of Education, Culture, Sports, Science and Technology (MEXT), and Japan Agency for Medical Research and development (AMED). M. K. was supported by JSPS KAKENHI Grant-in-Aid for Scientific Research on Innovative Areas (21121002 and 26119005). Y. M. was supported by JSPS KAKENHI Grants-in-Aid for Young Scientists (B) (23770111 and 25840021) and a Grant-in-Aid for Scientific Research (C) (15K06966). M. T. was supported by a JSPS KAKENHI Grant-in-Aid for Young Scientists (B) (23770109), a Grant-in-Aid for Scientific Research (C) (25440018), and a grant from the Kurata Memorial Hitachi Science and Technology Foundation. We would like to thank Dr. Tugarinov, of the University of Maryland, for providing the MSG gene and his advice regarding MSG protein production.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
TT. was an employee of SAIL Technologies, Inc. during the present research, and M.K. is a Research Advisor for the company.
Additional information
Yohei Miyanoiri and Yojiro Ishida have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Miyanoiri, Y., Ishida, Y., Takeda, M. et al. Highly efficient residue-selective labeling with isotope-labeled Ile, Leu, and Val using a new auxotrophic E. coli strain. J Biomol NMR 65, 109–119 (2016). https://doi.org/10.1007/s10858-016-0042-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-016-0042-0