Abstract
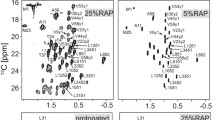
Magic-angle spinning (MAS) solid-state NMR (SSNMR) spectroscopy of uniformly-13C,15N labeled protein samples provides insight into atomic-resolution chemistry and structure. Data collection efficiency has advanced remarkably in the last decade; however, the study of larger proteins is still challenged by relatively low resolution in comparison to solution NMR. In this study, we present a systematic analysis of SSNMR protein spectra acquired at 11.7, 17.6 and 21.1 Tesla (1H frequencies of 500, 750, and 900 MHz). For two protein systems—GB1, a 6 kDa nanocrystalline protein and DsbA, a 21 kDa nanocrystalline protein—line narrowing is demonstrated in all spectral regions with increasing field. Resolution enhancement is greatest in the aliphatic region, including methine, methylene and methyl sites. The resolution for GB1 increases markedly as a function of field, and for DsbA, resolution in the C–C region increases by 42%, according to the number of peaks that can be uniquely picked and integrated in the 900 MHz spectra when compared to the 500 MHz spectra. Additionally, chemical exchange is uniquely observed in the highest field spectra for at least two isoleucine Cδ1 sites in DsbA. These results further illustrate the benefits of high-field MAS SSNMR spectroscopy for protein structural studies.







Similar content being viewed by others
References
Baldus M (2002) Correlation experiments for assignment and structure elucidation of immobilized polypeptides under magic angle spinning. Prog Nucl Magn Reson Spectrosc 41:1–47
Bennett AE, Rienstra CM, Auger M, Lakshmi KV, Griffin RG (1995) Heteronuclear decoupling in rotating solids. J Chem Phys 103:6951–6958
Bockmann A, Lange A, Galinier A, Luca S, Giraud N, Juy M, Heise H, Montserret R, Penin F, Baldus M (2003) Solid state NMR sequential resonance assignments and conformational analysis of the 2 × 10.4 kDa dimeric form of the Bacillus subtilis protein Crh. J Biomol NMR 27:323–339
Castellani F, van Rossum B, Diehl A, Schubert M, Rehbein K, Oschkinat H (2002) Structure of a protein determined by solid-state magic-angle spinning NMR spectroscopy. Nature 420:98–102
Cornilescu G, Delaglio F, Bax A (1999) Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR 13:289–302
Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A (1995) NMRPipe—a multidimensional spectral processing system based on Unix Pipes. J Biomol NMR 6:277–293
Detken A, Hardy EH, Ernst M, Meier BH (2002) Simple and efficient decoupling in magic-angle spinning solid-state NMR: the XiX scheme. Chem Phys Lett 356:298–304
Dillmann B, Elbayed K, Zeiger H, Weingertner MC, Plotto M, Engelke F (2007) A novel low-E field coil to minimize heating of biological samples in solid-state multinuclear NMR experiments. J Magn Reson 187:10–18
Doty FD, Kulkarni J, Turner C, Entzminger G, Bielecki A (2006) Using a cross-coil to reduce RF heating by an order of magnitude in triple-resonance multinuclear MAS at high fields. J Magn Reson 182:239–253
Franks WT, Zhou DH, Wylie BJ, Money BG, Graesser DT, Frericks HL, Sahota G, Rienstra CM (2005) Magic-angle spinning solid-state NMR spectroscopy of the beta1 immunoglobulin binding domain of protein G (GB1): 15N and 13C chemical shift assignments and conformational analysis. J Am Chem Soc 127:12291–12305
Fung BM, Khitrin AK, Ermolaev K (2000) An improved broadband decoupling sequence for liquid crystals and solids. J Magn Reson 142:97–101
Goddard TD, Kneller DG (2006) Sparky 3. University of California, San Francisco
Gronenborn AM, Filpula DR, Essig NZ, Achari A, Whitlow M, Wingfield PT, Clore GM (1991) A novel, highly stable fold of the immunoglobulin binding domain of streptococcal protein-G. Science 253:657–661
Guddat LW, Bardwell JCA, Martin JL (1998) Crystal structures of reduced and oxidized DsbA: investigation of domain motion and thiolate stabilization. Structure 6:757–767
Hediger S, Meier BH, Kurur ND, Bodenhausen G, Ernst RR (1994) NMR cross-polarization by adiabatic passage through the Hartmann-Hahn condition (APHH). Chem Phys Lett 223:283–288
Hong M, Jakes K (1999) Selective and extensive 13C labeling of a membrane protein for solid-state NMR investigations. J Biomol NMR 14:71–74
Igumenova TI, McDermott AE, Zilm KW, Martin RW, Paulson EK, Wand AJ (2004a) Assignments of carbon NMR resonances for microcrystalline ubiquitin. J Am Chem Soc 126:6720–6727
Igumenova TI, Wand AJ, McDermott AE (2004b) Assignment of the backbone resonances for microcrystalline ubiquitin. J Am Chem Soc 126:5323–5331
LeMaster DM, Kushlan DM (1996) Dynamical mapping of E. coli thioredoxin via 13C NMR relaxation analysis. J Am Chem Soc 118:9255–9264
Marley J, Lu M, Bracken C (2001) A method for efficient isotopic labeling of recombinant proteins. J Biomol NMR 20:71–75
Martin JL, Bardwell JCA, Kuriyan J (1993) Crystal structure of the DsbA protein required for disulphide bond formation in vivo. Nature 365:464–468
McDermott AE (2004) Structural and dynamic studies of proteins by solid-state NMR spectroscopy: rapid movement forward. Curr Opin Struct Biol 14:554–561
McDermott A, Polenova T, Bockmann A, Zilm KW, Paulson EK, Martin RW, Montelione GT (2000) Partial NMR assignments for uniformly (13C, 15N)-enriched BPTI in the solid state. J Biomol NMR 16:209–219
Morcombe CR, Zilm KW (2003) Chemical shift referencing in MAS solid state NMR. J Magn Reson 162:479–486
Morcombe CR, Gaponenko V, Byrd RA, Zilm KW (2004) Diluting abundant spins by isotope edited radio frequency field assisted diffusion. J Am Chem Soc 126:7196–7197
Oldfield E (2002) Chemical shifts in amino acids, peptides, and proteins: from quantum chemistry to drug design. Ann Rev Phys Chem 53:349–378
Opella SJ, Marassi FM (2004) Structure determination of membrane proteins by NMR spectroscopy. Chem Rev 104:3587–3606
Pauli J, Baldus M, van Rossum B, de Groot H, Oschkinat H (2001) Backbone and side-chain 13C and 15N resonance assignments of the alpha-spectrin SH3 domain by magic angle spinning solid state NMR at 17.6 Tesla. ChemBioChem 2:101–110
Schirra HJ, Renner C, Czisch M, Huber-Wunderlich M, Holak TA, Glockshuber R (1998) Structure of reduced DsbA from Escherichia coli in solution. Biochemistry 37:6263–6276
Smith ME, van Eck ERH (1999) Recent advances in experimental solid state NMR methodology for half-integer spin quadrupolar nuclei. Prog Nucl Magn Reson Spectrosc 34:159–201
Stringer JA, Bronnimann CE, Mullen CG, Zhou DH, Stellfox SA, Li Y, Williams EH, Rienstra CM (2005) Reduction of RF-induced sample heating with a scroll coil resonator structure for solid-state NMR probes. J Magn Reson 173:40–48
Takegoshi K, Nakamura S, Terao T (2001) 13C–1H dipolar-assisted rotational resonance in magic-angle spinning NMR. Chem Phys Lett 344:631–637
Wishart DS, Sykes BD (1994) The 13C chemical-shift index: a simple method for the identification of protein secondary structure using 13C chemical-shift data. J Biomol NMR 4:171–180
Acknowledgments
The authors thank the National Institute of Heath for funding through NIGMS (GM073770), NIGMS/Roadmap Initiative (GM075937) and Molecular Biophysics Training Grant (to LJS and AJN), David Hoyt, Jesse Sears, and Paul Ellis at the Environmental Molecular Science Laboratory (a national scientific user facility sponsored by the Department of Energy Office of Biological and Environmental Research) located at Pacific Northwest National Laboratory and operated for DOE by Batelle for their assistance in acquiring the 900 MHz data, Dr. Donghua Zhou for pulse sequence code, Dr. Trent Franks and Benjamin Fisher of the VOICE NMR Facility for technical assistance.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sperling, L.J., Nieuwkoop, A.J., Lipton, A.S. et al. High resolution NMR spectroscopy of nanocrystalline proteins at ultra-high magnetic field. J Biomol NMR 46, 149–155 (2010). https://doi.org/10.1007/s10858-009-9389-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-009-9389-9