Abstract
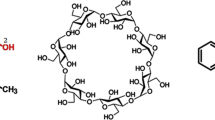
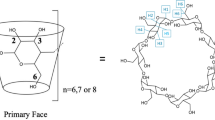
In our study, molecular docking and molecular dynamics (MD) simulations were performed in order to explore the interactions between human insulin and β-cyclodextrin (β-CD). Molecular docking study was performed using the Autodock v4.2 program to determine the number of β-CD molecules that adhere to the binding sites of insulin. A random structure docking approach using an initial ratio of 1:1 insulin-β-CD was conducted and from these, additional β-CDs were added. Molecular docking results revealed that a maximum of four β-CDs are able to bind to the insulin structure with the 1:3 insulin-β-CD ratio producing the lowest binding free energy. The docked conformations showed that hydrophobic interactions played a crucial role in insulin-β-CD conformational stability in addition to the formation of hydrogen bonds. A 50 ns MD simulation was further conducted using an NPT ensemble to verify the results obtained by molecular docking. The analysis of the MD simulation results of the 1:3 insulin-β-CD formation system conclude that a good interaction exists between insulin and β-CDs and the RMSD value obtained was 4.00 ± 0.50 Å. The RMSF profiles of insulin in the 1:3 insulin-β-CD formation also show reduced amino acid residues flexibility as compared to the free insulin system. The theoretical results indicated the presence of significant interactions between insulin and β-CD which could provide interesting insights into an insulin formulation.










Similar content being viewed by others
References
Williams, R., Gaal, L.V., Lucioni, C.: Assessing the impact of complications on the costs of Type II diabetes. Diabetologia 45, 13–17 (2002)
Ramachandran, A., Snehalatha, C., Shetty, A.S., Nanditha, A.: Trends in prevalence of diabetes in Asian countries. World J. Diabetes 3, 110–117 (2012)
Lu, S.Y., Jiang, Y.J., Jing, L.V., Wua, T.X., Yua, Q.S., Zhu, W.L.: Molecular docking and molecular dynamics simulation studies of GPR40 receptor-agonist interactions. J. Mol. Graph. Model. 28, 766–774 (2010)
Zhang, X., Qi, J., Lu, Y., He, W., Li, X., Wu, W.: Biotinylated liposomes as potential carriers for the oral delivery of insulin. Nanomedicine 10, 167–176 (2014)
Ullmann’s Encyclopedia of Industrial Chemistry 6th edn. Wiley-VCH, Chichester (2002)
Olsen, H.B., Ludvigsen, S., Kaarsholm, N.C.: Solution structure of an engineered insulin monomer at neutral pH. Biochemistry 35, 8836–8845 (1996)
Loftsson, T., Jarho, P., Másson, M., Järvinen, T.: Cyclodextrins in drug delivery. Expert. Opin. Drug Deliv. 2, 335–351 (2005)
Dodson, G., Steiner, D.: The role of assembly in insulin’s biosynthesis. Curr. Opin. Struck. Biol. 8, 189–194 (1998)
Dunn, M.F.: Zinc–ligand interactions modulate assembly and stability of the insulin hexamer—a review. Biometals 18, 295–303 (2005)
Szejtli, J.: Introduction and general overview of cyclodextrin chemistry. Chem. Rev. 98, 1743–1753 (1998)
Xiong, X.Y., Li, Q.H., Li, Y.P., Guo, L., Li, Z.L., Gong, Y.C.: Pluronic P85/poly(lactic acid) vesicles as novel carrier for oral insulin delivery. Colloids Surf. B 111, 282–288 (2013)
Elsayed, A.M.: Oral Delivery of Insulin: Novel Approaches, pp. 281–314. InTechOpen, Taif (2012)
Timmy, S.A., Victor, S.P., Sharma, C.P., Kumari, V.J.: Beta cyclodextrin complexed insulin loaded alginate microspheres—oral delivery system. Trends Biomater. Artif. Organs. 15, 48–53 (2002)
Ashada, H., Douen, T., Mizokoshi, Y., Fujita, T., Murakami, M., Yamamoto, A., Muranishi, S.: Absorption characteristics of chemically modified insulin derivatives with various fatty acids in the small and large intestine. J. Pharm. Sci. 84, 682–687 (1995)
Liu, H., Tang, R., Pan, W.S., Zhang, Y., Liu, H.: Potential utility of various protease inhibitors for improving the intestinal absorption of insulin in rats. J. Pharm. Pharmacol. 55, 1523–1529 (2003)
Salamat-Miller, N., Johnston, T.P.: Current strategies used to enhance the paracellular transport of therapeutic polypeptides across the intestinal epithelium. Int. J. Pharm. 294, 201–216 (2005)
Nakamura, K., Murray, R.J., Joseph, J.I., Peppas, N.A., Morishita, M., Lowman, A.M.: Oral insulin delivery using P(MAA-g-EG) hydrogels: effects of network morphology on insulin delivery characteristics. J. Cont. Release. 95, 589–599 (2014)
Choudhari, K.B., Labhasetwar, V.: Liposomes as a carrier for oral administration of insulin: effect of formulation factors. J. Microencapsul. 11, 319–325 (1994)
Al-Achi, A., Greenwood, R.: Erythrocytes as oral delivery systems for human insulin. Drug Dev. Ind. Pharm. 24, 67–72 (1998)
Damge, C., Vranckx, H., Balschmidt, P., Couvreur, P.: Poly (alkyl cyanoacrylate) nanospheres for oral administration of insulin. J. Pharm. Sci. 86, 1403–1409 (1997)
Chung, H., Kim, J., Um, J.Y., Kwon, I.C., Jeong, S.Y.: Self-assembled nanocubicle as a carrier for peroral insulin delivery. Diabetologia 45, 448–451 (2004)
Hua, Q., Weiss, M.A.: Comparative 2D NMR studies of human insulin and des-pentapeptide insulin: sequential resonance assignment and implications for protein dynamics and receptor recognition. Biochemistry 30, 5505–5515 (1991)
Abbaspour, A., Noori, A.: A cyclodextrin host-guest recognition approach to an electrochemical sensor for simultaneous quantification of serotonin and dopamine. Biosens. Bioelectron. 26, 4674–4680 (2011)
Franco, C., Schwingel, L., Lula, L., Sinisterra, R.D., Koeester, L., Bassani, V.L.: Studies on coumestro/β-cyclodextrin association : Inclusion complex characterization. Int. J. Pharm. 369, 5–11 (2009)
McEvoy, K., McMahon, H.E.M.: Antiprion action of new cyclodextrin analogues. Biochim. Biophys. Acta 1790, 1382–1386 (2009)
Aachmann, F.L., Otzen, D.E., Larsen, K.L., Wimmer, R.: Structural background of cyclodextrin-protein interactions. Prot. Eng. 16, 905–912 (2003)
Sajeesh, S., Sharma, C.P.: Cyclodextrin-insulin complex encapsulated polymethacrylic acid based nanoparticles for oral insulin delivery. Int. J. Pharm. 325, 147–154 (2006)
Krauland, A.H., Alonso, M.J.: Chitosan/cyclodextrin nanoparticles as macromolecular drug delivery system. Int. J. Pharm. 340, 134–142 (2007)
Yoshida, A., Yamamoto, M., Irie, T., Hirayama, F., Uekama, K.: Some pharmaceutical properties of 3-hydroxypropyl- and 2,3-dihydroxypropyl-β-cyclodextrins and their solubilizing and stabilizing abilities. Chem. Pharm. Bull. 37, 1059–1063 (1989)
WHO Toxicological evaluation of certain food additives and contaminants. β-Cyclodextrin, vol. 32, pp 173–193. World Health Organization, Geneva. WHO Food Additives Series (1993)
Fromming, K.H., Szejtli, J.: Pharmacokinetics and toxicology of cyclodextrins. In: Fromming, K.H., Szejtli, J. (eds.) Cyclodextrins in Pharmacy, pp. 33–45. Springer-Science+Bussines Media, London (1994)
Bellringer, M.E., Smith, T.G., Read, R., Gopinath, C., Olivier, P.: β-Cyclodextrin: 52 week toxicity studies in the rat and dog. Food Chem. Toxicol. 33, 367–376 (1995)
Hadaruga, D.I., Bals, D., Hadaruga, N.G.: Insulin-containing amino acids and oligopeptides/β-cyclodextrin supramolecular systems: molecular modeling and docking experiments. Chem. Bull. 54, 108–113 (2009)
Zoete, V., Meuwly, M., Karplus, M.: Investigation of glucose binding sites on insulin. Proteins 55, 568–581 (2004)
Berhanu, W.M., Masunov, A.E.: Controlling the aggregation and rate of release in order to improve insulin formulation : molecular dynamics study of full-length insulin amyloid oligomer models. J. Mol. Model. 18, 1129–1142 (2012)
Alexander, J.M., Clark, J.L., Brett, T.J., Stezowski, J.J.: β-cyclodextrin N-acetyl-l-phenylalanine clathrate dodecahydrate. Proc. Nat. Acad. Sci. 99, 5115 (2002)
Frisch, M.J., Trucks, G.W., Schlegel, H.B., Scuseria, G.E., Robb, M.A., Cheeseman, J.R., Montgomery Jr, J.A., Vreven, T., Kudin, K.N., Burant, J.C., Millam, J.M., Iyengar, S.S., Tomasi, J., Ba-rone, V., Mennucci, B., Cossi, M., Scalmani, G., Rega, N., Pet-ersson, G.A., Nakatsuji, H., Hada, M., Ehara, M., Toyota, K., Fukuda, R., Hasegawa, J., Ishida, M., Nakajima, T., Honda, Y., Kitao, O., Nakai, H., Klene, M., Li, X., Knox, J.E., Hratchian, H.P., Cross, J.B., Bakken, V., Adamo, C., Jaramillo, J., Gom-perts, R., Stratmann, R.E., Yazyev, O., Austin, A.J., Cammi, R., Pomelli, C., Ochterski, J.W., Ayala, P.Y., Morokuma, K., Voth, G.A., Salvador, P., Dannenberg, J.J., Zakrzewski, V.G., Dap-prich, S., Daniels, A.D., Strain, M.C., Farkas, O., Malick, D.K., Rabuck, A.D., Raghavachari, K., Foresman, J.B., Ortiz, J.V., Cui, Q., Baboul, A.G., Clifford, S., Cioslowski, J., Stefanov, B.B., Liu, G., Liashenko, A., Piskorz, P., Komaromi, I., Martin, R.L., Fox, D.J., Keith, T., Al-Laham, M.A., Peng, C.Y., Nanayakkara, A., Challacombe, M., Gill, P.M.W., Johnson, B., Chen, W., Wong, M.W., Gonzalez, C., Pople, J.A.: Gaussian 03, Revision D. 01. Gaussian, Pittsburgh (2004)
Morris, G.M., Huey, R., Lindstrom, W., Sanner, M.F., Belew, R.K., Goodsell, D.S., Olson, A.J.: Autodock4 and AutoDockTools4: automated docking with selective receptor flexiblity. J. Comput. Chem. 16, 2785–2791 (2009)
Morris, G.M., Goodsell, D.S., Halliday, R.S., Huey, R., Hart, W.E., Belew, R.K., Olson, A.J.: Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 19, 1639–1662 (1998)
Laskowski, R.A., Swindells, M.B.: LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 51, 2778–2786 (2011)
van Der Spoel, D., Lindahl, E., Hess, B., Groenhof, G., Mark, A.E., Berendsen, H.J.C.: GROMACS: fast, flexible, and free. J. Comput. Chem. 26, 1701–1718 (2005)
Hess, B., Kutzner, C., van der Spoel, D., Lindahl, E.: GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 4, 435–447 (2008)
Stocker, U., van Gunsteren, W.F.: Molecular dynamics simulation of hen egg white lysozyme: a test of the GROMOS96 force field against nuclear magnetic resonance data. Proteins Struct. Funct. Bioinform. 40, 145–153 (2000)
Rivail, L., Chipot, C., Maigret, B., Bestel, I., Sicsic, S., Tarek, M.: Large-scale molecular dynamics of a G protein-coupled receptor, the human 5-HT4 serotonin receptor, in a lipid bilayer. J. Mol. Struct. 817, 19–26 (2007)
Abraham, M.J., Gready, J.E.: Optimization of parameters for molecular dynamics simulation using smooth particle-mesh Ewald in GROMACS 4.5. J. Comput. Chem. 32, 2031–2040 (2011)
Hess, B., Bekker, H., Berendsen, H.J.C., Fraaije, J.G.E., Fraaije, J.G.E.M.: LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 18, 1463–1472 (1997)
Chandler, D.: Review article interfaces and the driving force of hydrophobic assembly. Nature 437, 640–647 (2005)
Cooper, A.: Effect of cyclodextrins on the thermal stability of globular proteins. J. Am. Chem. Soc. 114, 9208–9209 (1992)
Berman, H.M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T.N., Weissig, H., Shindyalov, I.N., Bourne, P.E.: The protein data bank. Nucleic Acids Res. 28, 235–242 (2000)
Mikami, B., Sato, M., Shibata, T., Hirose, M., Aibara, S., Katsube, Y., Morita, Y.: Three-dimensional structure of soybean beta-amylase determined at 3.0 Å resolution: preliminary chain tracing of the complex with alpha-cyclodextrin. J. Biochem. 112, 541–546 (1992)
Yokota, T., Tonozuka, T., Shimura, Y., Ichiwa, K., Kamitori, S., Sakano, Y.: Structures of Thermoactinomyces vulgaris R-47 alpha-amylase II complexed with substrate analogues. Biosci. Biotechnol. Biochem. 65, 619–626 (2001)
Pullen, R.A., Lindsay, D.G., Wood, S.P., Tickle, I.J., Blundell, T.L., Wollmer, A., Krail, G., Brandenburg, D., Zahn, H., Gliemann, J., Gammeltoft, S.: Receptor-binding region of insulin. Nature 259, 369–373 (1976)
Pittman, I.T., Tager, H.S.: A spectroscopic investigation of the conformational dynamics of insulin in solution. Biochemistry 34, 10578–10590 (1995)
Hua, Q., Weiss, M.A.: Mechanism of insulin fibrillation. The structure of insulin under amyloidogenic conditions resembles a protein-folding. J. Biol. Chem. 279, 21449–21460 (2004)
Zhang, R., Wang, Z., Ling, B., Liu, Y., Liu, C.: Docking and molecular dynamics studies on the interaction of four imidazoline derivatives with potassium ion channel (Kir6.2). Mol. Simul. 36, 166–174 (2010)
Falconi, M., Cambria, M.T., Cambria, A., Desideri, A.: Structure and stability of the insulin dimer investigated by molecular dynamics simulation. J. Biomol. Struct. Dyn. 18, 761–772 (2001)
Acknowledgments
The study was financially supported by the funding from Universiti Sains Malaysia through the Research University Grant (Grant No. 1001/PKIMIA/815099). The authors gratefully acknowledge the technical assistance from the staff of the Department of Chemistry, Faculty of Science, Universiti Putra Malaysia.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest
Rights and permissions
About this article
Cite this article
Muhammad, E.F., Adnan, R., Latif, M.A.M. et al. Theoretical investigation on insulin dimer-β-cyclodextrin interactions using docking and molecular dynamics simulation. J Incl Phenom Macrocycl Chem 84, 1–10 (2016). https://doi.org/10.1007/s10847-015-0576-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10847-015-0576-x